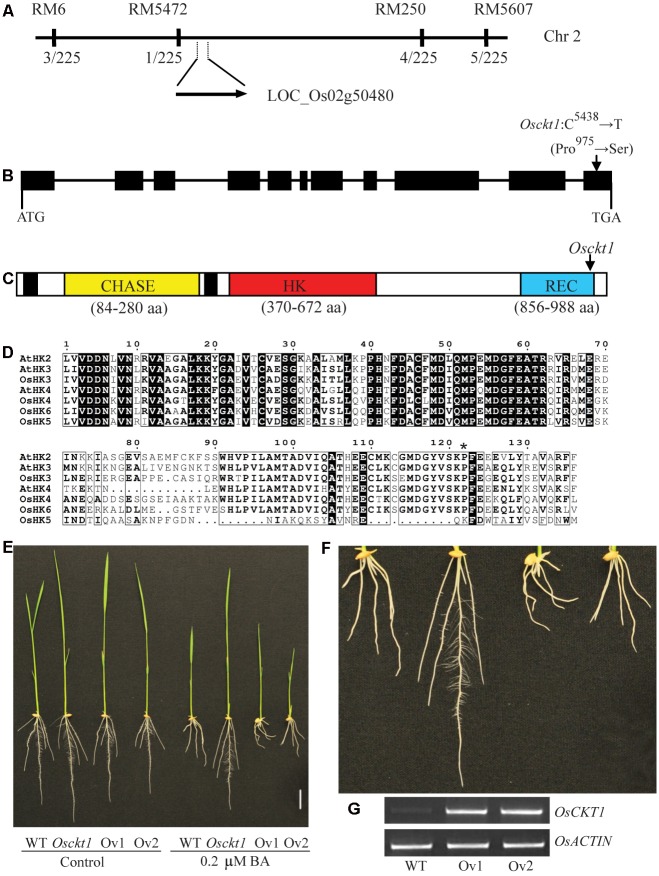
FIGURE 2.
Map-based cloning of OsCKT1. (A) OsCKT1 was mapped to a 2137 kb region on chromosome 2. The rates of recombinants in the F2 population are listed below the molecular markers. (B) Gene structure of OsCKT1. Black boxes represent exons, and lines indicate introns and the 5′ and 3′ untranslated regions. The point mutation in the last exon is indicated. (C) Secondary structure analysis of OsCKT1 using the CDD database (https://www.ncbi.nlm.nih.gov/cdd). Black boxes represent transmembrane domains. The point mutation results in a substitution of Proline by Serine in the REC domain. (D) Protein sequence alignment of the REC domain of HKs from Arabidopsis and rice. Identical and highly similar amino acids are highlighted in black and framed in black by ESPript, respectively. The mutated residue in Osckt1 was indicated by asterisk. (E–G) Complementation analysis of the Osckt1 mutant. Two independent lines of over-expression transgenic plants (Ov1 and Ov2) in the Osckt1 mutant background were displayed (E) and the enlarged view of roots under BA treatment was shown (F). RT-PCR analysis (G) of OsCKT1 in roots of Ov1 and Ov2. Bar = 2 cm.