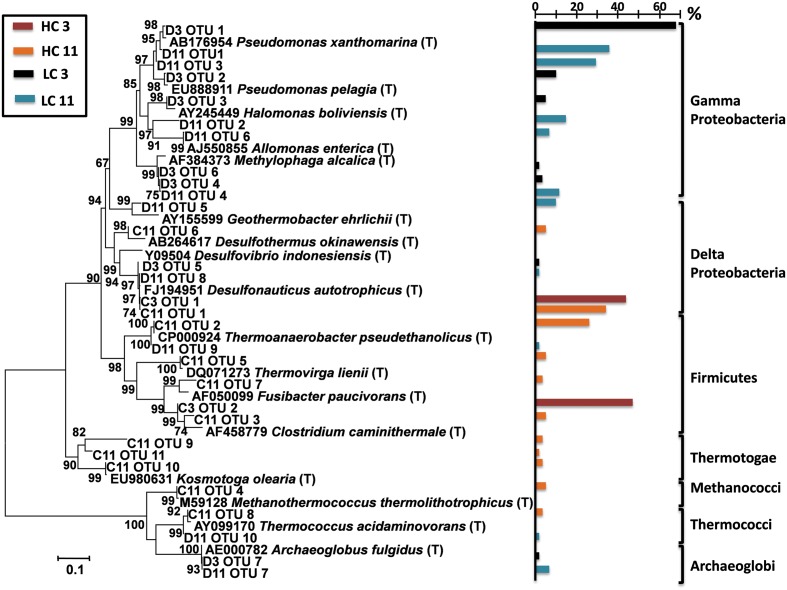
FIGURE 1.
Phylogenetic analysis of full length 16S rRNA genes extracted from metagenomic data using EMIRGE (Miller, 2013). Operational Taxonomic Units (OTUs) were compared to the RDP database, and the most closely related 16S rRNA genes from type strains (designated with a T) were used for phylogenetic analysis of metagenomic OTUs. A bootstrapped (5,000 replicates) Neighbor-Joining tree is shown. The colored bars indicated the proportion of 16S reads that were assembled into respective full length 16S sequences for each of the detected phylotypes (percentage scale shown in scale bar above). Bacterial and archaeal divisions/families are indicated and color coded. D3 = LC3; D11 = LC11; C3 = HC3; and C11 = HC11.