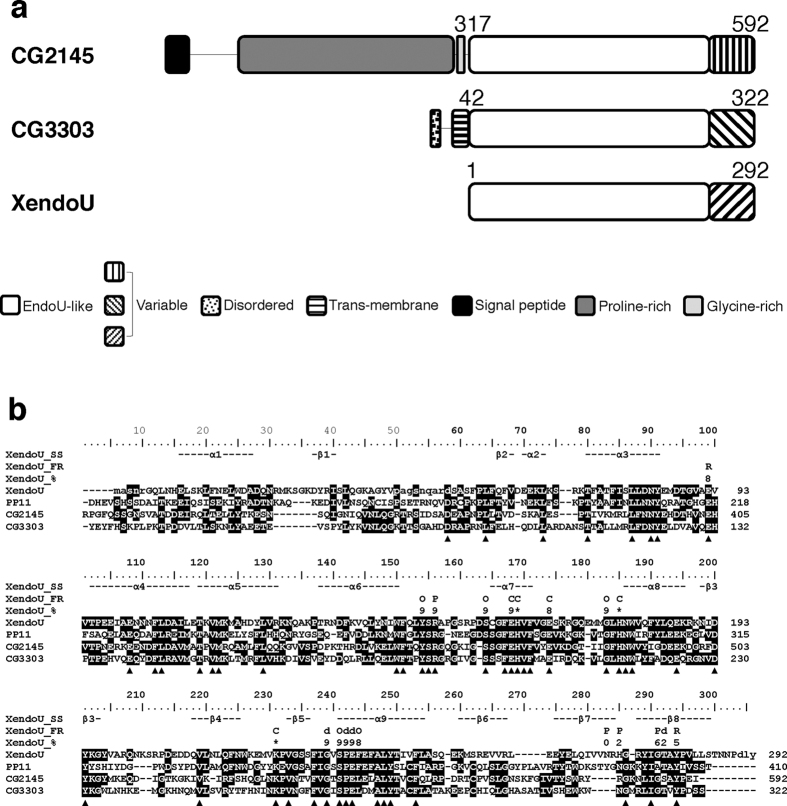
Figure 1. Sequence analyses of Drosophila CG2145 and CG3303 gene products.
(a) Scheme of CG2145 and CG3303 compared to XendoU. Sequence numbering indicates starting and ending residues of XendoU and the CG2145 and CG3303 regions homologous to XendoU (see panel b). These regions comprise a highly conserved segment (EndoU-like, white boxes) and a variable segment (striped boxes). The N-terminal regions of CG2145 and CG3303 comprise additional segments not present in XendoU. The N-terminal region of CG2145 is significantly longer and comprises a signal peptide (black box), a long disordered and proline-rich region (gray box), and a short glycine-rich segment (light gray box). The N-terminal region of CG3303 comprises a disordered segment (dotted box) and a predicted trans-membrane region (horizontally-striped box). (b) Multiple sequence alignment of CG3303 and CG2145 products with human PP11 and X. laevis XendoU. Residues that are not visible in the experimental XendoU structure are lower-case. Identical residues at the same alignment position have a black background. Positions where all residues are identical are indicated by black triangles. XendoU_SS: secondary structure elements detected in the 3D XendoU structure; α-helices and β-strands are indicated with α and β and a number indicating their progression along the amino acid sequence. XendoU_FR: XendoU residues (i) with a key functional role, i.e., those reported to diminish (d) or abolish (C) catalysis if mutated; affect RNA binding (R); in contact with the phosphate molecule in XendoU 3D structure (P); and (ii) shown to interact with XendoU key-residues in the 3D structure (O). XendoU_%: conservation of XendoU residues in a multiple sequence alignment comprising over 700 family members (see Supplementary Table 1); numbers 0, 1, 2, …, 8, 9 indicate conservation in the ranges: <10%, 10–19%, 20–29%, …, 80–89%, 90–99%, respectively; catalytic residues, which are 100% conserved in the extended multiple sequence alignment, are indicated by∗.