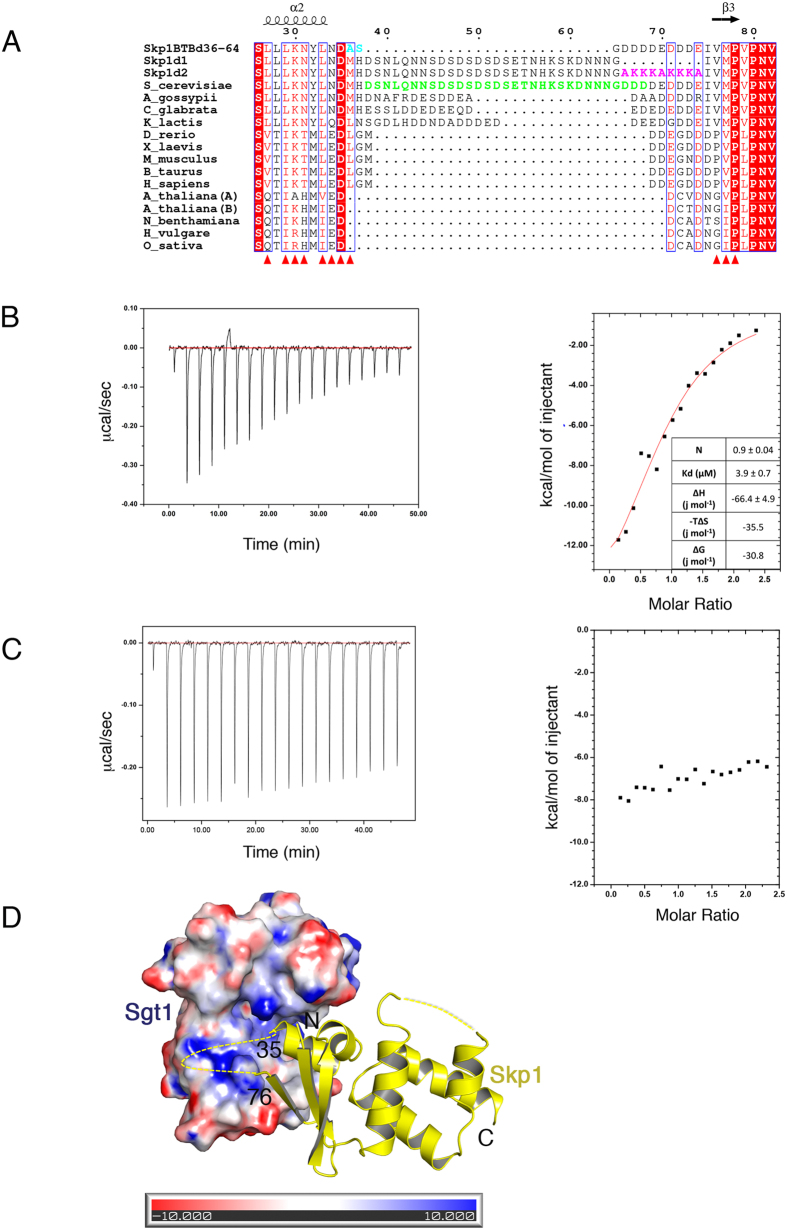
Figure 4. Contribution of a Charged Sequence to Sgt1-Skp1 Association.
(A) Sequence alignment of Skp1 homologs including the Skp1BTB∆ (Skp1BTBd36–64) construct used for crystallization, Skp1∆1 (Skp1d1) and Skp1∆2 (Skp1d2) mutants. S. cerevisiae Skp1 has a loop (green) that is not conserved in higher eukaryotes. The loop is replaced by an Ala-Ser linker (cyan) in Skp1BTB∆. The mutated residues in Skp1∆2 are coloured magenta. White text on a red background indicates strict identity, red text indicates similarity in a group, a blue frame indicates similarity across groups. Residues that contribute to the interface between Sgt1 and Skp1 are shown as red triangles. Generated using ESPript56. (B) Left: Raw ITC data for the interaction between Sgt1 (13 μM dimer) and Skp1Δ1 (130 μM) at 25 °C. Right: Dilution-corrected and integrated heats for the ITC data are fitted according to a single binding site model assuming dimeric Sgt1. (C) As Figure B but for the interaction between Sgt1 (12 μM dimer) and Skp1∆2 (145 μM) at 25 °C. (D) The complex of Sgt1TPR (electrostatic surface potential ± 10 kT/e), generated using PDB2PQR and APBS57,58 and Skp1BTB∆ (shown in cartoon). The unconserved deleted loop (residues 36–64) and following acidic stretch (residues 65–73) are shown as a dotted line.