Fig. 2.
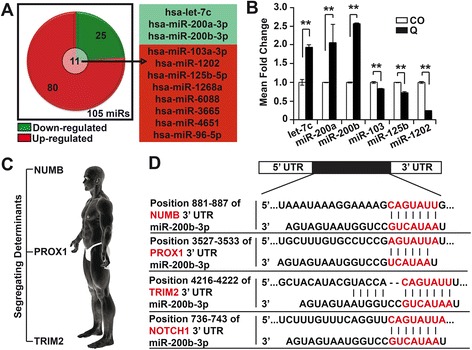
MiR-200b-3p could be a cell fate determinant miRNA. a In silico analyses was used to identify 11 out of 105 differentially regulated miRs, as players in Notch signaling/cell fate determination. b Using qRT-PCR, the microarray data was validated by Taqman® miRNA assay. RNA isolation from untreated (CO) and treated (Q, 50 μM) AsPC1 cells, followed by reverse transcription and PCR. RNU44 was used to normalize the expression levels and the average fold change of the CO was set to 1, while the expression level in the quercetin-treated cells was related to the control. c Human homolog of the segregating determinants. d Putative binding sites of miR-200b-3p in the 3’UTR of the segregating determinants (Numb, Prox1, and Trim2) and Notch1 was identified using TargetScan database. The seed-binding regions are marked in red. All assays were performed in triplicates, repeated at least three times and the means ± S.D. are shown. *P < 0.05, **P < 0.01
