Fig. 6.
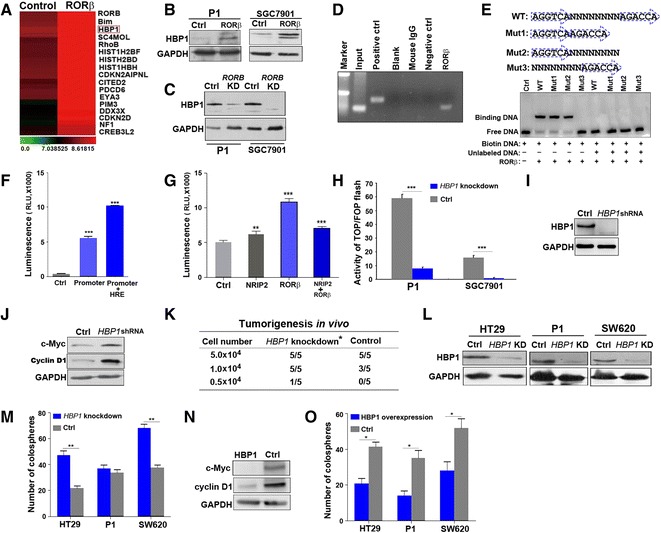
HBP1 is a crucial target of RORβ in regulation of the Wnt pathway. a Altered mRNA in RORβ-overexpressing cells. SGC7901 cells were transiently transfected with RORB/pReceiver and control pReceiver plasmids for 24 h. Total RNA was purified for global cDNA GeneChip scanning. The most significant up-regulated genes are listed. b Detection of HBP1 in RORβ-overexpressing cells. HBP1 was detected by western blotting in cells overexpressing RORβ. P1 cells infected with blank lentivirus and SGC7901 cells transfected with pReceiver plasmids were used as controls c Detection of HBP1 in RORB-knockdown cells. HBP1 was detected by western blotting in RORB-knockdown cells that were produced by infection with RORB shRNA lentivirus. HBP1 expression was significantly decreased after knockdown of RORB. P1 and SGC7901 cells infected with scrambled shRNA lentivirus were used as controls. d ChIP analysis of the interaction between RORβ and HBP1 upstream DNA. DNA fragments were immunoprecipitated by anti-myc-tag antibodies agarose in RORβ-overexpressing SGC7901 cells after sonication. PCR was used for the detection of the HBP1 upstream DNA sequence. The results showed that RORβ bound with the region upstream of HBP1 DNA. Blank, normal mouse IgG was used as a negative control, and anti-RNA polymerase II was used as a positive control. e RORβ binds to hormone response elements (HRE) upstream of the HBP1 promoter region. An EMSA assay was used to identify the seed region for RORβ binding within upstream hormone response elements of the HBP1 promoter region. Three mutants containing different potential binding sequences were constructed. The results showed that the hormone response element sequence AGGTCA is essential for RORβ binding with the HBP1 promoter region. f HRE increased the activity of the promoter. Plasmids containing HRE or the promoter of HBP1 were co-transfected into 239 T cells for 24 h. Luciferase activity was evaluated by the dual-luciferase reporter assay system. The results showed that HRE increased the HBP1 promoter activity, ***p < 0.001 (ANOVA). pRL3 plasmids were used as a control. g NRIP2 attenuated RORβ transactivation. The luciferase activity of the HBP1 promoter was determined in 293 T cells at 24 h after co-transfection of RORB and/or NRIP2 as well as in pRL3 plasmids containing HRE and the HBP1 promoter. The results showed that NRIP2 attenuated RORβ transactivation, ***p < 0.001 (ANOVA). Blank pRL3 plasmids were used as a control. h NRIP2 could not activate Wnt activity in HBP1-silenced cells. Wnt activity was evaluated by a luciferase activity assay in HBP1-silenced cells and scrambled P1 and SGC7901 cells (control) 24 h after co-transfection with Top/Fop flash reporters and NRIP2 plasmids. The results showed that NRIP2 could not activate Wnt activity in cells after silencing HBP1. ***p < 0.001 (ANOVA). i Detection of HBP1 in HBP1-knockdown cells. HBP1 was detected by western blotting in the SGC7901 cells with knockdown of HBP1 by shRNAs. SGC7901 cells transfected with scrambled shRNAs as a control. j Wnt activity in HBP1-knockdown cells. c-Myc and cyclin D1 were detected by western blotting in the above HBP1-knockdown and scrambled SGC7901 cells. k Tumorigenicity of HBP1-knockdown cells. HBP1-knockdown SGC7901 cells and their control cells infected with blank lentivirus were injected into naked Balb/c mice, respectively. Tumor formation was quantified within 4 weeks. The results showed that silence of HBP1 significantly increased tumorigenicity (p < 0.05, Multivariate logistic analysis). l HBP1 expression in HBP1-knockdown colorectal cancer cells. Colorectal cancer cells were infected with HBP1 shRNA lentivirus for knockdown of HBP1. HBP1 was detected by western blotting in these HBP1-knockdown and scrambled colorectal cancer cells (control). m Quantification of colospheres in HBP1-knockdown cells. Colospheres were counted in HBP1-knockdown and scrambled cells. The number of colospheres was significantly increased in HBP1-knockdown cells. **p < 0.01 (ANOVA). n Detection of the Wnt downstream targets in HBP1-overexpressing cells. c-Myc and cyclin D1 were analyzed by western blotting in HBP1-overexpressing and control (transfected with pCMV-XL4 plasmids) P1 cells. o Quantification of colospheres in HBP1-overexpressing cells. The number of colospheres was counted in the above HBP1-overexpressing and control cells. The results showed that HBP1 significantly inhibited colosphere formation. *p < 0.05 (ANOVA)
