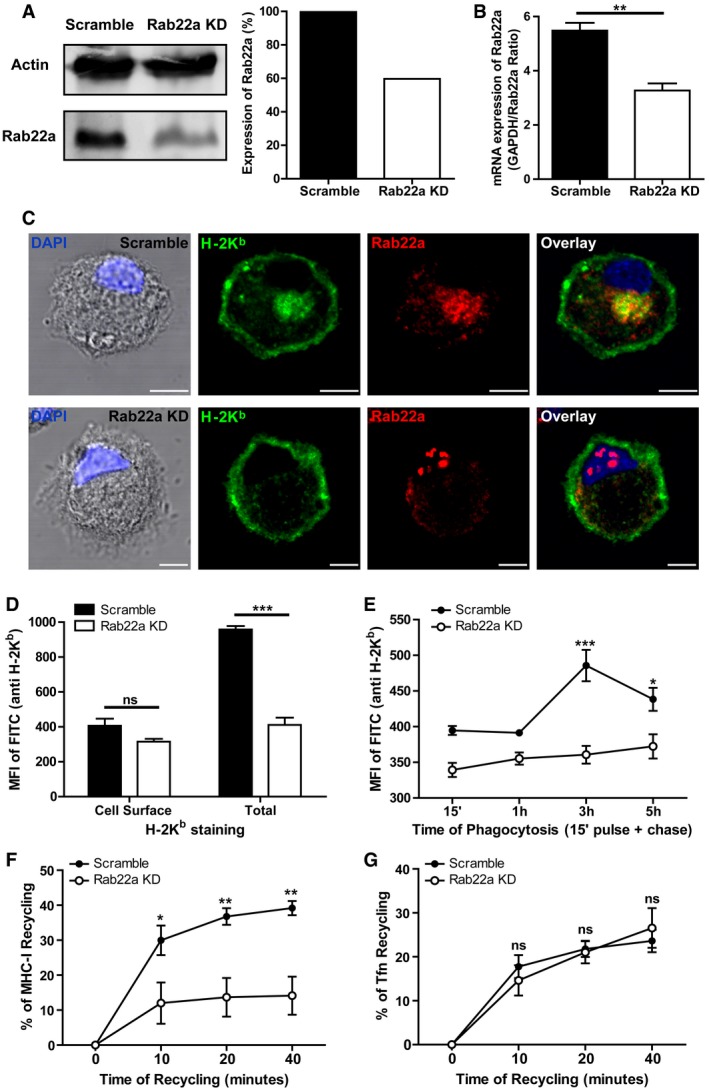
Immunoblotting and densitometry quantification of Rab22a in JAWS‐II DCs infected with lentiviruses encoding a random sequence (Scramble) and a shRNA specific for silencing Rab22a (Rab22a KD). Data are representative of at least three independent experiments.
RNA extraction and qPCR quantification of Rab22a in the same cells analyzed in (A). Data show mean ± SEM of triplicate values and are representative of two independent experiments. **P = 0.0047.
IF labeling and confocal microscopy analysis showing the distribution of MHC‐I molecules (H‐2Kb, green) and Rab22a (red) in Scramble and Rab22a KD JAWS‐II DCs. Nuclei stained with DAPI and DIC images are shown on the left. Overlay is shown in the right panels. Scale bars: 5 μm. Data are representative of at least 30 images analyzed for each experimental condition from three independent experiments.
FACS analysis of MHC‐I labeled in intact (cell surface) and permeabilized (total) Scramble and Rab22a KD JAWS‐II cells. Data represent mean ± SEM of triplicate values and are representative of three independent experiments. P = 0.1082 (ns) and ***P = 0.0003.
MHC‐I staining on isolated phagosomes was measured by FACS at the indicated time periods after 3‐μm LB internalization by Scramble and Rab22a KD JAWS‐II DCs. Data represent mean ± SEM of three independent experiments. ***P < 0.001 at 3 h and *P < 0.05 at 5 h between both DC types.
MHC‐I molecules recycling ability was measured by FACS at the indicated time periods by Scramble and Rab22a KD JAWS‐II DCs. Data represent mean ± SEM of three independent experiments. *P < 0.05 at 10 min and **P < 0.01 at 20 and 40 min.
The transferrin (Tfn) recycling ability was measured by FACS at the indicated time periods by Scramble and Rab22a KD JAWS‐II DCs. Data represent mean ± SEM of three independent experiments. P > 0.05 (ns) at 10, 20, and 40 min.
‐test was performed. In (E–G), a two‐way ANOVA and the Bonferroni post‐test were performed.