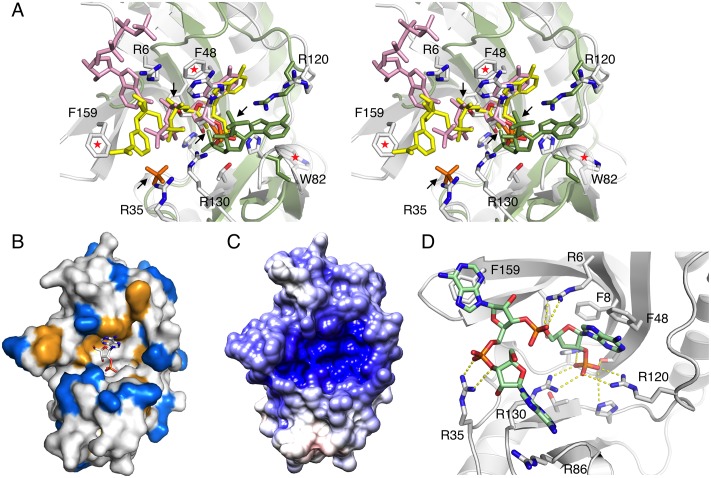
Fig 7. Evidence for an RNA binding surface from individual structures of nucleotide complexes.
A. Superimposed are the complexes of LigT (white) with 2′-AMP (white), NADP+ (yellow), ATP (pink), and phosphate (orange), as well as the complex of a viral RNase L agonist [29] with adenosine 2′,5′-bisphosphate (green). All the ligands have a phosphate group in the active site, and superposition reveals other nearby features for recognizing phosphate or aromatic base groups in RNA. Arrows indicate the different phosphate binding sites, while red asterisks denote aromatic rings potentially involved in stacking of RNA bases. B. Distribution of aromatic (orange) and basic (blue) groups in LigT. The catalytic site is indicated by the bound reaction product 2′-AMP. C. Surface electrostatic potential of LigT. The catalytic groove has a high positive charge potential. D. Model of binding for a 3-residue RNA molecule with a 2′-phospho group in the active site. Yellow dashed lines indicate the polar contacts by the phosphate moieties. Also pay attention to stacking of bases against Phe8, Phe159, and Arg86. Note Arg120 has flipped over to interact with the active-site phosphate, when the model was energy-minimized and subjected to a short MD simulation.