Abstract
Objective
This study aims to explore the expression pattern and prognostic significance of miR-33a in non-small cell lung cancer (NSCLC) treated with adjuvant chemotherapy.
Methods
MiR-33aexpression in NSCLC was analyzed in silico using the GEO database and was subsequently confirmed by quantitative RT-PCR in 147 NSCLC biopsies. Among these, 32 of these biopsies were paired with adjacent non-neoplastic tissues. The survival analysis of NSCLC by Kaplan-Meier estimates was stratified based on miR-33a expression. In addition, multivariate survival analysis in corresponding groups of NSCLC patients was conducted by Cox proportional hazards regression model.
Results
The in silico analysis of miR-33a expression in NSCLC resulted to its down-regulation in different tumor types. The expression level of miR-33a was lower in each grade of NSCLC tumor biopsies than in normal lung tissues. Univariate and multivariate survival analysis further established that low miR-33a expression was an important risk factor for overall survival and disease free survival in NSCLC patients.
Conclusion
Our study implied that miR-33a expression levels may have an essential role in NSCLC progression, and could act as a specific and sensitive biomarker for NSCLC patients who have undergone adjuvant chemotherapy.
Introduction
Lung cancer is the third most frequently diagnosed cancer and the leading cause of cancer-related mortality worldwide. There are approximately 1.8 million new lung cancer cases annually [1]. Patients with non-small cell lung cancer (NSCLC), which accounts for approximately 75–80% of the total lung cancer incidents, are mostly diagnosed at the advanced stages of the disease [2, 3]. For NSCLC patients who have undergone surgical resection, the American Society of Clinical Oncology (ASCO) guidelines recommend the use of post-operative therapeutic strategies including adjuvant external radiation therapy or cytotoxic chemotherapy combined with molecular targeted therapy [2, 4–7]. However, current staging approaches are inadequate in predicting and diagnosing the outcome of NSCLC treatments due to the unavailability of potential biomarkers for molecular targeted or personalized treatments. Therefore, improvement in molecular genetics diagnosis and the prediction of prognosis for targeted treatments and clinical decisions are urgently required.
MicroRNAs (miRNAs) are a large number of small noncoding RNA genes found to be aberrantly expressed in various types of malignancies and function either as oncogenes or tumor suppressors. This implies that miRNAs play a vital role in tumorigenesis and cancer progression [8–11]. Furthermore, these have also been shown to be involved in oncogenesis mechanisms, which can serve as potential cancer biomarkers [12]. Therefore, the understanding of miRNA expression patterns as potential biomarkers for the diagnosis and prognosis of personalized targeted therapies and clinical decision and management has just started to unfold [12–15]. The identification of a miRNA signature that can predict the benefit from adjuvant chemotherapy would be definitely helpful for the clinical decision and management of NSCLC in patients. However, it remains ambiguous whether the miRNA signature can predict the clinical decisions of NSCLC, including major adjuvant chemotherapy or TNM stage.
Although a majority of patients are diagnosed initially by imaging techniques, the prognosis of patients with this gene mutation remains poor in traditional treatment. Fluorodeoxyglucose (FDG)-PET/CT scans play an important role not only in the staging of lung cancer, but also in predicting and assessing treatment responses at the present time. However, these could not provide genetic information useful for predicting adjuvant chemotherapeutic or gene targeting therapeutic options. The use of genomics-based diagnosis for patients with resectable tumors and the combination of chemotherapy, radiation therapy in conjunction with molecular targeted therapy, would improve overall survival (OS) and disease-free survival (DFS) in patients with locally advanced lung cancer. These miRNAs have the potential to regulate the expression of thousands of corresponding target genes, and are able to govern a comprehensive range of biological functions such as cellular proliferation, differentiation, apoptosis, immune response, and the maintenance of cell and tissue identity [16, 17]. The results of molecular exploration may improve clinical decisions and management for NSCLC patients [18]. Advances in genomics, transcriptomics and proteomics have resulted in the generation of many candidate biomarkers with potential clinical significance.
There has previously reported that low levels of miR-33a expression were found in NSCLC patients in clinical and suggested that the miR-33 family might play a significant role in NSCLC prognosis and patient survival [19]. However, few are known on the associations between miR-33 levels and NSCLC patients treated with adjuvant chemotherapy. Therefore, in order to investigate whether specific and sensitive biomarkers can predict the clinical outcome of NSCLC at the molecular level, including the prognosis and response to adjuvant chemotherapy, we focused on the role of miRNAs, and attempted to determine a link between its expression and NSCLC survival.
Material and Methods
Ethics statement
The study was approved by the Ethics Committee of Shanghai Tenth People’s Hospital, Tongji University School of Medicine (SHSY-IEC-pap-15-18). Each participant provided a written informed consent before participating in this study. All specimens were handled and made anonymous according to ethical and legal standards.
GEO data acquisition and processing
MiR-33 expression in NSCLC biopsies was analyzed in silico using the GEO database. The GEO database provides a multimodal data repository and retrieval system for high-throughput functional genomic data generated by microarray and next-generation sequencing technologies and can be acquired from the GEO website (The Gene Expression Omnibus, http://www.ncbi.nlm.nih.gov/geo/) [20, 21].
Acquisition of clinical specimens
Fresh frozen tissue samples from NSCLC patients, who underwent surgical resection between 2008 and 2012, were obtained from the tissue bank of Shanghai Tenth People’s Hospital. These samples included paired tumor and adjacent non-cancerous tissues (n = 32), as well as a large cohort of individual NSCLC biopsies (n = 115). The histological typing of these tumors was performed according to the World Health Organization criteria. Staging was performed according to the Seventh Edition of the American Joint Commission on Cancer (AJCC) tumor-node-metastasis (TNM) staging system for NSCLC [22], and patient data was collected up to May 30, 2015. The clinical information recorded included the patient’s characteristics, tumor characteristics, OS, DFS and chemotherapy status.
RNA isolation and the detection of miR-33a expression by qRT-PCR
Total RNA was extracted from NSCLC and normal tissues using TRIZOL reagent, according to manufacturer’s instructions. RNA concentration was measured by a spectrophotometer, and the quality of all RNA samples was assessed by electrophoresis on 1.5% denaturing agarose gels. qRT-PCR was carried out using a Taqman miRNA PCR kit (Applied Biosystems, Foster City, CA) according to manufacturer’s instructions. Briefly, total RNA was reverse-transcribed to cDNA using AMV reverse transcriptase (TaKaRa, Dalian, China) and the stem- loop RT primers (Applied Biosystems). Real-time PCR was performed using TaqMan miRNA probes on the Applied Biosystems 7300 Sequence Detection System (Applied Biosystems). U6 was used as the internal control. The 2-δδCT method was used to quantify the expression levels of miR-33a, and the expression status (e.g., high levels or low expression levels) was recorded.
Statistical analysis
All statistical analyses were accomplished with IBM SPSS statistics software Version 20.0 for Windows. The expression of miR-33a was represented as the mean ± standard deviation. Independent t-test was used to examine the differences between two groups and Chi-square test was used to evaluate the differences in rates between groups. Kaplan-Meier curves were used to determine the OS of various groups, and results were compared using the log-rank test. Univariate and multivariate survival analyses were based on the Cox regression model, and this model was used to identify the independent factors that had significant effects on survival. A P-value < 0.05 was considered statistically significant.
Results
Analysis of miR-33a expression in cancer using the in silico data platform
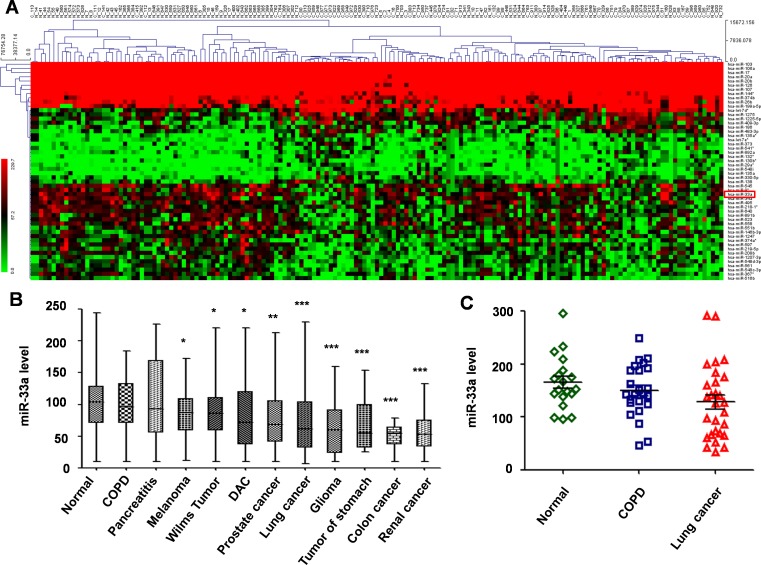
Microarray description and raw data have been made available in the GEO database with reference number GSE59153. This project analyzed the peripheral blood profiles of patients from various cancers (diseases), and controls. These included normal controls (n = 94), lung cancer individuals (n = 71) and patients (n = 940) have been screened for complete miRNA (1,049 miRNAs) repertoire, according to miRNAse V12-14. Each miRNA was measured in seven replicates at least, and the median of the replica was computed. The in silico data sample hierarchal clustering of the gene expression microarrays data was performed using the MEV 4.7.1 clustering software. The 52 miRNAs with significant changes (miR-33a included, P < 0.001, Fold change (FC) ≥ 2 or ≤ 0.5) were filtered out, and following the expression of differentially expressed miRNAs was assessed with reference to the prognosis of the NSCLC patient (Fig 1A). Among the 52 miRNAs from the GEO database, 19 miRNAs were up-regulated and 33 miRNAs were down-regulated. This included previously published up-regulated miRNAs in lung cancer, such as miR-130b*, miR-135a and miR-138, and down-regulated miRNAs, such as miR-126, miR-144, miR-20a/b and miR-218-1 [19].
Fig 1. Analysis of miR-33a expression from GEO datasets.
A, Data from the GEO dataset (GSE59153) was clustered using MEV 4.7.1 software. B, Relative expression levels of miR-33a in different cancers vs. normal controls from the GEO dataset (GSE59153). C, Relative expression levels of miR-33a in NSCLC and adjacent normal tissues from patients with cancerous and non-cancerous lung diseases from the GEO dataset (GSE24709).
After ruling out these known differentially expressed miRNAs in NSCLC, the rest of the differentially expressed miRNAs in normal, lung cancer, chronic obstructive pulmonary disease (COPD), and pancreatitis with different types of tumors (disease) were further analyzed; and miR-33a was the only one found to be significantly down-regulated in all the different tumor types (Fig 1B).
In addition, we also downloaded peripheral profiles from patients with cancerous and non-cancerous lung diseases (including 19 normal controls, 28 lung cancer patients and 24 COPD samples) (GSE24709), and found that also miR-33a was significantly reduced in lung cancer (P = 0.035; Fig 1C).
However, we noticed that miR-33a was reported to highly express in tumor samples from glioma, gastric, renal and colon cancer [23–25]. Thus, in our current study, we were particularly interested in miR-33a expression and clinical significance to NSCLC prognosis.
Analysis of miR-33a expression in NSCLCs and normal lung tissues by qRT-PCR
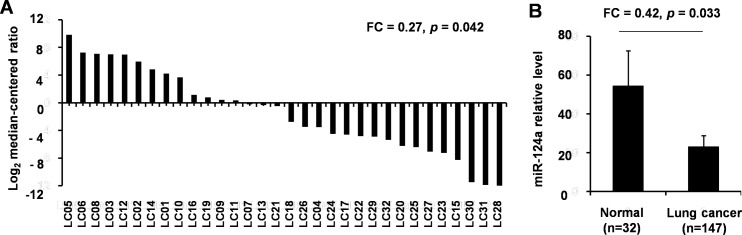
In order to further confirm the results of the in silico gene microarray study, we analyzed the miR-33a expression in large samples from clinic. A total of 147 (94 grade I-II, 52 grade III-IV) tumor specimens and 32 normal lung specimens were included for this analysis. The 32 samples had paired tumor and adjacent non-cancerous tissues (n = 32). The miR-33a analysis in these paired samples by qRT-PCR demonstrated that its expression levels were significantly lower in NSCLC tumor biopsies relative to adjacent non-neoplastic tissues. This difference was statistically significant (P = 0.042, FC = 0.27; Fig 2A). Moreover, the analysis of other tumor specimens also revealed that the level of miR-33a expression was significantly lower in each grade of NSCLC tumor biopsy than in normal lung tissues (P = 0.033, FC = 0.42; Fig 2B).
Fig 2. Expression of miR-33a in normal lung and NSCLC tissues.
A, The expression levels (log2 median-centered ratio) of miR-33a in 32 paired cancers vs. adjacent non-tumor tissues were analyzed. B, The miR-33a expression in NSCLC samples (n = 147) and paired adjacent non-tumor tissues (n = 32) was analyzed.
Correlation of miR-33 expression with demographic factors and clinical characteristics
In order to validate whether miR-33a expression levels were affected by clinical characteristics, we explored the correlation of miR-33a expression with demographic and clinical factors. All 147 patients of NSCLC included in this study demonstrated that miR-33a expression levels were negatively correlated with lymph-node metastasis (P = 0.028), TNM stage (P = 0.038), invasion of the lung membrane (P = 0.039) and the diameter of the tumor (P = 0.027) as shown in Table 1. However, we did not observe any association between miR-33a expression and patient gender, age, smoking history, tumor differentiation, histology, vascular invasion and adjuvant chemotherapy (P > 0.05, Table 1).
Table 1. Univariate analysis of overall survival in NSCLC patients stratified based on clinical characteristics.
| Factor | Variable | N | miR-33a expression (Median) | P value | Overall survival | |
|---|---|---|---|---|---|---|
| Months (Median) | 95% CI (Mean) | |||||
| Age | ≥ 60 | 92 | 29.24 | 0.121 | 32.26 | 29.32–35.21 |
| < 60 | 55 | 18.75 | 32.02 | 29.12–34.93 | ||
| Gender | Male | 89 | 23.1 | 0.928 | 32.11 | 29.27–34.94 |
| Female | 58 | 22.09 | 35.87 | 33.01–38.74 | ||
| Smoking history | Never | 35 | 24.01 | 0.118 | 32.05 | 28.74–29.16 |
| Ever | 41 | 19.18 | 30.25 | 27.43–35.06 | ||
| Unknown | 71 | 26.54 | 30.88 | 25.39–33.48 | ||
| Lymph-node metastasis | Negative | 85 | 27.06 | 0.028 | 31.28 | 24.72–33.87 |
| Positive | 50 | 14.14 | 29.34 | 26.43–32.58 | ||
| Unknown | 12 | 18.73 | 30.26 | 28.34–33.16 | ||
| Tumor differentiation | Poorly | 51 | 18.96 | 0.051 | 28.93 | 26.54–30.65 |
| Moderately | 88 | 25.43 | 29.33 | 27.06–31.54 | ||
| Well | 8 | 26.08 | 31.69 | 29.64–33.71 | ||
| Histology | Adenocarcinoma | 94 | 24.63 | 0.654 | 28.43 | 26.55–31.29 |
| Squamous cell carcinoma | 52 | 26.07 | 32.38 | 28.43–35.66 | ||
| TNM stage | I-II | 94 | 28.09 | 0.038 | 33.58 | 30.19–35.48 |
| III-IV | 52 | 22.14 | 27.68 | 24.36–29.13 | ||
| Invasion of lung membrane | Negative | 30 | 23.35 | 0.039 | 32.67 | 29.89–34.26 |
| Positive | 105 | 16.75 | 28.03 | 26.74–30.52 | ||
| Unknown | 12 | 18.94 | 29.31 | 27.65–35.43 | ||
| Vascular invasion | Negative | 132 | 23.16 | 0.652 | 30.39 | 28.45–32.33 |
| Positive | 9 | 19.03 | 29.04 | 26.71–31.31 | ||
| Unknown | 6 | 20.36 | 28.33 | 26.58–31.46 | ||
| Chemotherapy | Negative | 74 | 22.35 | 0.557 | 26.94 | 23.06–28.94 |
| Positive | 62 | 21.03 | 32.47 | 29.64–34.53 | ||
| Unknown | 11 | 23.07 | 28.03 | 26.54–32.19 | ||
| Diameter | ≥ 5 cm | 37 | 17.43 | 0.027 | 24.86 | 25.39–30.01 |
| < 5 cm | 110 | 26.03 | 32.43 | 31.21–34.68 | ||
Univariate analysis of OS in NSCLC patients stratified based on clinical characteristics
A univariate survival analysis was performed through Kaplan-Meier estimates by stratifying NSCLC patients based on clinical factors (including gender, age, smoking history, lymph-node metastasis, tumor differentiation, histology, TNM stage, invasion of the lung membrane, vascular invasion, and tumor size). Median follow-up was 39.6 months (range: 14.6 to 89.6 months). Results of the univariate analyses are shown in Table 1.
As expected, there was a significant association between shorter OS and classical prognostic factors such as lymph-node metastasis (P = 0.045), TNM stage (P = 0.012), invasion of the lung membrane (P = 0.018), and tumor size (≥ 5 cm, P< 0.001). In addition, the univariate analysis using the Cox proportional hazards regression model revealed that lymph node metastasis (P = 0.04, HR = 1.54 [1.16, 1.86]), TNM stage (P = 0.01, HR = 2.29 [2.72, 2.38]), invasion of the lung membrane (P = 0.02, HR = 1.76 [1.45, 1.99]) and diameter (P = 0.001, HR = 2.56 [2.03, 3.45]) were positively correlated with poorer prognosis (Table 2).
Table 2. Cox regression model analysis for prognosis based on various clinical characteristics of NSCLC patients.
| Factor | HR | 95% CI (univariate) | P value | miR-33a multivariate analysis | ||
|---|---|---|---|---|---|---|
| HR | 95% CI (multivariate) | P value | ||||
| Age | 0.95 | 0.71–1.15 | 0.78 | |||
| Gender | 0.88 | 0.54–1.26 | 0.36 | |||
| Smoking history | 1.22 | 0.73–1.38 | 0.12 | |||
| Lymph-node metastasis | 1.54 | 1.16–1.86 | 0.04 | 1.78 | 1.20–2.66 | 0.02 |
| Tumor differentiation | 0.78 | 0.54–1.11 | 0.16 | |||
| Histology | 1.19 | 0.88–1.24 | 0.11 | |||
| TNM stage | 2.29 | 2.72–3.38 | 0.01 | 2.43 | 1.86–2.98 | 0.008 |
| Invasion of lung membrane | 1.76 | 1.45–1.99 | 0.02 | |||
| Vascular invasion | 0.91 | 0.68–1.24 | 0.75 | |||
| Diameter (≥ 5 cm) | 2.56 | 2.03–3.45 | 0.001 | 3.17 | 2.26–5.03 | <0.001 |
| Low miR-33a expression | 1.66 | 1.21–2.24 | 0.028 | |||
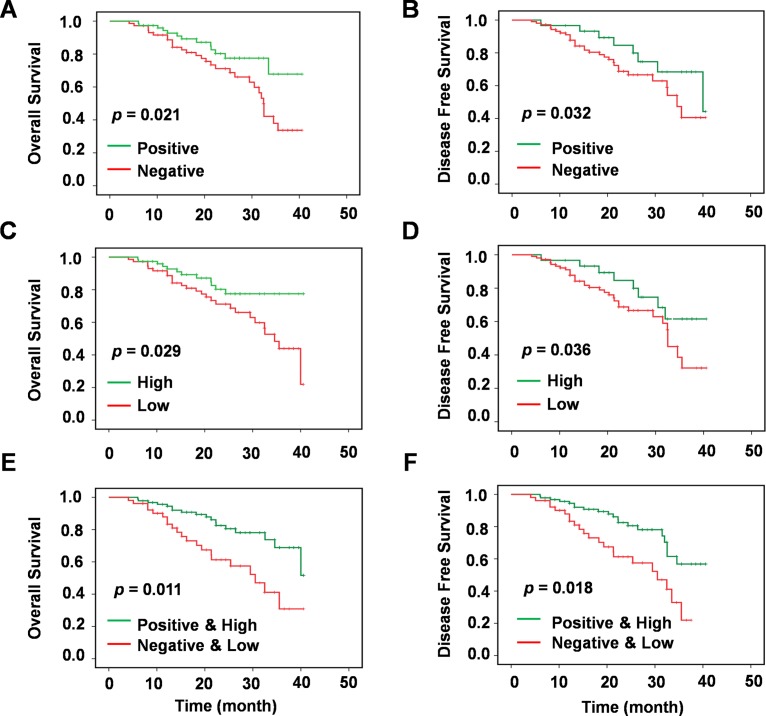
In order to assess the influence of chemotherapy on the prognosis of NSCLC patients, Kaplan-Meier survival curves were plotted and log rank analysis was performed. The results revealed that adjuvant chemotherapy was significantly associated with increase OS (P = 0.021, Fig 3A) and DFS (P = 0.032, Fig 3B) in NSCLC patients.
Fig 3. Survival analysis of miR-33a expression and chemotherapy in NSCLCs.
Univariate survival analysis of overall survival and disease-free survival in lung carcinoma as determined by Kaplan-Meier plots estimates based on chemotherapy (Negative vs. Positive) in (A) and (B), miR-33a expression (Low vs. High) in (C) and (D), and miR-33a expression and chemotherapy (non-chemotherapy and low miR-33a expression vs. chemotherapy and high miR-33a expression) in (E) and (F), respectively.
Thus, significantly reduced OS was associated with patients who had not undergone adjuvant chemotherapy or had lymph node metastasis, invasion of the lung membrane, or increased tumor size (≥ 5 cm).
Assessment of miR-33a as a prognostic tumor-marker for NSCLC patient survival
Results of the survival analysis demonstrated that patients with low miR-33a expression levels had poor OS rates compared to patients with high miR-33a expression. The low expression of miR-33a was significantly associated with decreased OS (P = 0.029, Fig 3C) and DFS (P = 0.036, Fig 3D) in NSCLC patients. Similarly, the low expression of miR-33a was also positively correlated with poor prognosis (P = 0.028, HR = 1.66 [1.21, 2.24]) (Table 2).
In order to further assess the contribution of other variables with miR-33a as a prognostic marker in NSCLC patients, we also performed a multivariate survival analysis, using the Cox proportional hazards regression model. This analysis initially included all parameters that were predictive of OS in the univariate analysis of the entire study group, as presented in Table 2 (age, gender, smoking history, lymph-node metastasis, tumor differentiation, histology, vascular invasion and diameter, and invasion of the lung membrane). A forward stepwise procedure was adopted to obtain the final model of significant predictors for OS, which consist of factors including lymph-node metastasis, diameter, TNM stage, and expression of miR-33a. According to this analysis, low miR-33a expression was identified as a predictor of shorter OS in NSCLC patients (Table 2).
Analysis of the predictive value of miR-33a expression for adjuvant chemotherapy
As adjuvant chemotherapy provides primary treatment after surgical operations in majority of NSCLC cases, the OS and DFS of patients were consequently explored based on the treatment signature. Adjuvant chemotherapy was identified to be significantly associated with increased OS (P = 0.021, Fig 3A) and DFS (P = 0.032, Fig 3B) in patients in this cohort. The Kaplan-Meier univariate and multivariate Cox proportional hazards regression survival analysis was further conducted to determine whether adjuvant chemotherapy and/or miR-33a expression were associated with OS and DFS. When this adjuvant chemotherapy data was analyzed based on the expression of miR-33a, OS and DFS were both observed to be significantly longer in NSCLC patients with high miR-33a expression as opposed to patients who were untreated or with a low expression (P = 0.011 and 0.018, respectively) (Fig 3E and 3F), which suggests that NSCLC patients with lower miR-33a expression has poor prognosis even when treated with adjuvant chemotherapy.
Discussion
The identification of specific and sensitive tumor-markers for revealing human malignancies is urgently required to decrease the global morbidity and mortality rate caused by cancer [26, 27]. With the effort of identifying ideal cancer markers, qRT-PCR methodologies have been extensively explored [15, 28]. In this study, we have established an effective strategy that allowed us to identify miRNA-based biomarkers for NSCLC. NSCLC biopsies and adjacent non- cancerous tissues were assessed for miR-33a expression. The expression of miR-33a was observed to be down-regulated in NSCLC patients. In this large sample population, we observed that the elevated expression of miR-33a was associated with the better prognosis of NSCLC patients who have undergone cytotoxic chemotherapy; which was consistent with a previously published study. Gong et al.[29] stated that miR-33a was expressed at lower levels in metastatic NSCLC cells. In this study, qRT-PCR was conducted to measure miR-33a expression levels in 53 pairs of NSCLC tumor and non-tumor tissue samples, and results demonstrated that the low-expression of miR-33a was predictive of poor prognosis in NSCLC patients [29]. In another study, the use of miR-33a was validated as a novel therapeutic target for colon carcinoma in a mouse model of preclinical study [24]. Therefore, based on these results it is reasonable to indicate that miR-33a expression levels may play a critical role in NSCLC progression; which could develop as a promising, specific and sensitive diagnostic biomarker for NSCLC patients at advanced stages. This also implies that miR-33a may be a novel and valuable tumor-marker, and increasing the cellular levels of miR-33a may be a novel therapeutic strategy for the treatment of patients with advanced NSCLC.
In general, human cancers that are comparable other diseases, are easier to treat and control when revealed at the early stage of disease progression [30, 31]. Distant metastasis usually triggers more than 80% of cancer deaths and involves a complicated sequence of steps where cancer cells leave the original position and migrate to other segments of the body through the circulatory and lymphatic system [32, 33]. Qi et al. [34] revealed that miR-33a expression is very poor in extreme metastatic breast cancer cell lines than noncancerous breast epithelial cells and non-metastatic breast cancer cells. At the same time, miR-33a has also been shown to inhibit breast cancer cell growth, migration and invasion together with the suppression of in vivo tumor growth and the lung metastasis of breast cancer cells [34]. Keet al. [35] also described that human metastatic melanoma cells have low miR-33a/b expression and is involved in the regulation of in vivo functions by acting as a tumor suppressor through targeting HIF-1α. The identification of this miR-33a/HIF-1α axis has been proposed to be a novel approach for the management of melanoma. [35]. Tumor recurrence and metastasis is crucial in patients not only under treatment with traditional chemotherapies, but also with more current molecular targeted therapies [18, 36]. The identification of miRNA-based markers for various types of cancers could help molecular based cancer classification [28, 37]. The important role of these miRNAs have been shown to play a role in cancer by targeting various signaling pathways and therapeutic responses to the target gene has delivered a new opening for developing novel agents and methodologies [38].These above results revealed that miR-33a could be used as a prospective therapeutic RNA mimic for the treatment of patients with metastasis/advanced carcinoma. Thus, it is necessary to screen miRNAs in a genome-wide manner, and determine all differentially expressed miRNAs in NSCLC patients for the guiding clinical management and use of adjuvant chemotherapy. The emergence of the association between miR-33a expression and chemotherapy from our study suggests a promising role for this biomarker in response to chemotherapy.
However, not all published studies suggest a positive association between miR-33a levels and cancer-relative diagnosis and therapy. MiR-33a level has been shown to be negatively correlated with the target gene TWIST and was over-expressed in chemo-resistant osteosarcoma; which resulted in the down-regulation of TWIST, and increased osteosarcoma cell resistance to cisplatin [39]. It was suggested that inhibition of miR-33a/TWIST signaling could be a latent novel approach to develop neoadjuvant chemotherapy for osteosarcoma. Moreover, understanding the molecular features of lung malignancies would help in targeted therapy development.
Although contemporary findings on the link between NSCLCs and miRNAs have drastically stretched our realization on the signaling pathway and its association in the pathogenic developments of NSCLC, our understanding of underlying mechanisms that integrate the activity of the molecular pathway remains incomplete. An extensive search on the regulatory roles of miRNAs in the signaling pathway may contribute to the possible option of using miRNAs as predictive, specific and sensitive tumor-markers.
Conclusion
In conclusion, our data confirmed that with adjuvant chemotherapeutic treatment, the median OS and DFS of patients improved. Furthermore, the analyses firstly demonstrated that the high expression of miR-33a, together with adjuvant chemotherapy, greatly improved OS and DFS in NSCLC patients. Particularly, the application of miR-33a as specific and sensitive biomarkers may also be useful for predicting therapeutic responses in advanced NSCLC patients, which could lead to a superior level of personalized therapy. Thus, MiR-33a can be regarded as a potential tumor biomarker for chemosensitivity in NSCLC patients.
Acknowledgments
This study was supported in part by grants from the National Natural Science Foundation of China (81372175, 81301993, 81472202, and 81302065), Shanghai health and family planning commission projects (201540228 and 201440398) and Shanghai Natural Science Foundation (16ZR1428900).
Data Availability
All relevant data are within the paper.
Funding Statement
This study was supported in part by grants from the National Natural Science Foundation of China (81372175, 81301993, 81472202, and 81302065), Shanghai health and family planning commission projects (201540228 and 201440398) and Shanghai Natural Science Foundation (16ZR1428900).
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA: a cancer journal for clinicians. 2015;65(2):87–108. Epub 2015/02/06. [DOI] [PubMed] [Google Scholar]
- 2.Masters GA, Temin S, Azzoli CG, Giaccone G, Baker S Jr., Brahmer JR, et al. Systemic Therapy for Stage IV Non-Small-Cell Lung Cancer: American Society of Clinical Oncology Clinical Practice Guideline Update. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2015;33(30):3488–515. Epub 2015/09/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friboulet L, Olaussen KA, Pignon JP, Shepherd FA, Tsao MS, Graziano S, et al. ERCC1 isoform expression and DNA repair in non-small-cell lung cancer. The New England journal of medicine. 2013;368(12):1101–10. Epub 2013/03/22. PubMed Central PMCID: PMCPmc4054818. 10.1056/NEJMoa1214271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodrigues G, Choy H, Bradley J, Rosenzweig KE, Bogart J, Curran WJ Jr., et al. Adjuvant radiation therapy in locally advanced non-small cell lung cancer: Executive summary of an American Society for Radiation Oncology (ASTRO) evidence-based clinical practice guideline. Practical radiation oncology. 2015;5(3):149–55. Epub 2015/05/10. 10.1016/j.prro.2015.02.013 [DOI] [PubMed] [Google Scholar]
- 5.Ettinger DS, Wood DE, Akerley W, Bazhenova LA, Borghaei H, Camidge DR, et al. Non-small cell lung cancer, version 1.2015. Journal of the National Comprehensive Cancer Network: JNCCN. 2014;12(12):1738–61. Epub 2014/12/17. [DOI] [PubMed] [Google Scholar]
- 6.Reck M, Heigener DF, Mok T, Soria JC, Rabe KF. Management of non-small-cell lung cancer: recent developments. Lancet (London, England). 2013;382(9893):709–19. Epub 2013/08/27. [DOI] [PubMed] [Google Scholar]
- 7.Ramnath N, Dilling TJ, Harris LJ, Kim AW, Michaud GC, Balekian AA, et al. Treatment of stage III non-small cell lung cancer: Diagnosis and management of lung cancer, 3rd ed: American College of Chest Physicians evidence-based clinical practice guidelines. Chest. 2013;143(5 Suppl):e314S–40S. Epub 2013/05/10. 10.1378/chest.12-2360 [DOI] [PubMed] [Google Scholar]
- 8.Yates LA, Norbury CJ, Gilbert RJ. The long and short of microRNA. Cell. 2013;153(3):516–9. Epub 2013/04/30. 10.1016/j.cell.2013.04.003 [DOI] [PubMed] [Google Scholar]
- 9.Martello G, Rosato A, Ferrari F, Manfrin A, Cordenonsi M, Dupont S, et al. A MicroRNA targeting dicer for metastasis control. Cell. 2010;141(7):1195–207. Epub 2010/07/07. 10.1016/j.cell.2010.05.017 [DOI] [PubMed] [Google Scholar]
- 10.Wang R, Wang ZX, Yang JS, Pan X, De W, Chen LB. MicroRNA-451 functions as a tumor suppressor in human non-small cell lung cancer by targeting ras-related protein 14 (RAB14). Oncogene. 2011;30(23):2644–58. Epub 2011/03/02. 10.1038/onc.2010.642 [DOI] [PubMed] [Google Scholar]
- 11.Kim J, Zhang Y, Skalski M, Hayes J, Kefas B, Schiff D, et al. microRNA-148a is a prognostic oncomiR that targets MIG6 and BIM to regulate EGFR and apoptosis in glioblastoma. Cancer research. 2014;74(5):1541–53. Epub 2014/01/16. PubMed Central PMCID: PMCPmc3947487. 10.1158/0008-5472.CAN-13-1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu SL, Chen HY, Chang GC, Chen CY, Chen HW, Singh S, et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer cell. 2008;13(1):48–57. Epub 2008/01/03. 10.1016/j.ccr.2007.12.008 [DOI] [PubMed] [Google Scholar]
- 13.Kadara H, Behrens C, Yuan P, Solis L, Liu D, Gu X, et al. A five-gene and corresponding protein signature for stage-I lung adenocarcinoma prognosis. Clinical cancer research: an official journal of the American Association for Cancer Research. 2011;17(6):1490–501. PubMed Central PMCID: PMC3079395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen HY, Yu SL, Chen CH, Chang GC, Chen CY, Yuan A, et al. A five-gene signature and clinical outcome in non-small-cell lung cancer. The New England journal of medicine. 2007;356(1):11–20. 10.1056/NEJMoa060096 [DOI] [PubMed] [Google Scholar]
- 15.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell research. 2008;18(10):997–1006. Epub 2008/09/04. 10.1038/cr.2008.282 [DOI] [PubMed] [Google Scholar]
- 16.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. Epub 2004/01/28. [DOI] [PubMed] [Google Scholar]
- 17.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nature reviews Genetics. 2004;5(7):522–31. Epub 2004/06/24. 10.1038/nrg1379 [DOI] [PubMed] [Google Scholar]
- 18.Han Y, Jia C, Cong X, Yu F, Cai H, Fang S, et al. Increased Expression of TGFbetaR2 Is Associated with the Clinical Outcome of Non-Small Cell Lung Cancer Patients Treated with Chemotherapy. PloS one. 2015;10(8):e0134682 Epub 2015/08/08. PubMed Central PMCID: PMCPmc4529313. 10.1371/journal.pone.0134682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer cell. 2006;9(3):189–98. Epub 2006/03/15. 10.1016/j.ccr.2006.01.025 [DOI] [PubMed] [Google Scholar]
- 20.Database resources of the National Center for Biotechnology Information. Nucleic acids research. 2015. Epub 2015/11/29. [DOI] [PMC free article] [PubMed]
- 21.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic acids research. 2013;41(Database issue):D991–5. Epub 2012/11/30. PubMed Central PMCID: PMCPmc3531084. 10.1093/nar/gks1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Annals of surgical oncology. 2010;17(6):1471–4. Epub 2010/02/25. 10.1245/s10434-010-0985-4 [DOI] [PubMed] [Google Scholar]
- 23.Wang H, Sun T, Hu J, Zhang R, Rao Y, Wang S, et al. miR-33a promotes glioma-initiating cell self-renewal via PKA and NOTCH pathways. The Journal of clinical investigation. 2014;124(10):4489–502. Epub 2014/09/10. PubMed Central PMCID: PMCPmc4191031. 10.1172/JCI75284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ibrahim AF, Weirauch U, Thomas M, Grunweller A, Hartmann RK, Aigner A. MicroRNA replacement therapy for miR-145 and miR-33a is efficacious in a model of colon carcinoma. Cancer research. 2011;71(15):5214–24. Epub 2011/06/22. 10.1158/0008-5472.CAN-10-4645 [DOI] [PubMed] [Google Scholar]
- 25.Li X, Luo F, Li Q, Xu M, Feng D, Zhang G, et al. Identification of new aberrantly expressed miRNAs in intestinal-type gastric cancer and its clinical significance. Oncology reports. 2011;26(6):1431–9. Epub 2011/08/30. 10.3892/or.2011.1437 [DOI] [PubMed] [Google Scholar]
- 26.Zen K, Zhang CY. Circulating microRNAs: a novel class of biomarkers to diagnose and monitor human cancers. Medicinal research reviews. 2012;32(2):326–48. Epub 2012/03/03. 10.1002/med.20215 [DOI] [PubMed] [Google Scholar]
- 27.Harris L, Fritsche H, Mennel R, Norton L, Ravdin P, Taube S, et al. American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2007;25(33):5287–312. Epub 2007/10/24. [DOI] [PubMed] [Google Scholar]
- 28.Liu R, Zhang C, Hu Z, Li G, Wang C, Yang C, et al. A five-microRNA signature identified from genome-wide serum microRNA expression profiling serves as a fingerprint for gastric cancer diagnosis. European journal of cancer (Oxford, England: 1990). 2011;47(5):784–91. Epub 2010/11/30. [DOI] [PubMed] [Google Scholar]
- 29.Yang L, Yang J, Li J, Shen X, Le Y, Zhou C, et al. MircoRNA-33a inhibits epithelial-to-mesenchymal transition and metastasis and could be a prognostic marker in non-small cell lung cancer. Scientific reports. 2015;5:13677 Epub 2015/09/04. PubMed Central PMCID: PMCPmc4556976. 10.1038/srep13677 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Cykert S, Dilworth-Anderson P, Monroe MH, Walker P, McGuire FR, Corbie-Smith G, et al. Factors associated with decisions to undergo surgery among patients with newly diagnosed early-stage lung cancer. Jama. 2010;303(23):2368–76. Epub 2010/06/17. PubMed Central PMCID: PMCPmc4152904. 10.1001/jama.2010.793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klopfleisch R, Kohn B, Gruber AD. Mechanisms of tumour resistance against chemotherapeutic agents in veterinary oncology. Veterinary journal (London, England: 1997). 2015. Epub 2015/11/04. [DOI] [PubMed] [Google Scholar]
- 32.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331(6024):1559–64. Epub 2011/03/26. 10.1126/science.1203543 [DOI] [PubMed] [Google Scholar]
- 33.Herbst RS, Heymach JV, Lippman SM. Lung cancer. The New England journal of medicine. 2008;359(13):1367–80. Epub 2008/09/26. 10.1056/NEJMra0802714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang C, Zhang Y, Ding W, Lin Y, Huang Z, Luo Q. MiR-33a suppresses breast cancer cell proliferation and metastasis by targeting ADAM9 and ROS1. Protein & cell. 2015;6(12):881–9. Epub 2015/10/29. PubMed Central PMCID: PMCPmc4656205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou J, Xu D, Xie H, Tang J, Liu R, Li J, et al. miR-33a functions as a tumor suppressor in melanoma by targeting HIF-1alpha. Cancer biology & therapy. 2015;16(6):846–55. Epub 2015/04/22. PubMed Central PMCID: PMCPmc4622471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neoptolemos JP, Stocken DD, Friess H, Bassi C, Dunn JA, Hickey H, et al. A randomized trial of chemoradiotherapy and chemotherapy after resection of pancreatic cancer. The New England journal of medicine. 2004;350(12):1200–10. Epub 2004/03/19. 10.1056/NEJMoa032295 [DOI] [PubMed] [Google Scholar]
- 37.Camps C, Buffa FM, Colella S, Moore J, Sotiriou C, Sheldon H, et al. hsa-miR-210 Is induced by hypoxia and is an independent prognostic factor in breast cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2008;14(5):1340–8. Epub 2008/03/05. [DOI] [PubMed] [Google Scholar]
- 38.Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nature reviews Molecular cell biology. 2010;11(4):252–63. Epub 2010/03/11. 10.1038/nrm2868 [DOI] [PubMed] [Google Scholar]
- 39.Zhou Y, Huang Z, Wu S, Zang X, Liu M, Shi J. miR-33a is up-regulated in chemoresistant osteosarcoma and promotes osteosarcoma cell resistance to cisplatin by down-regulating TWIST. Journal of experimental & clinical cancer research: CR. 2014;33:12. Epub 2014/01/29. PubMed Central PMCID: PMCPmc3974149. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.