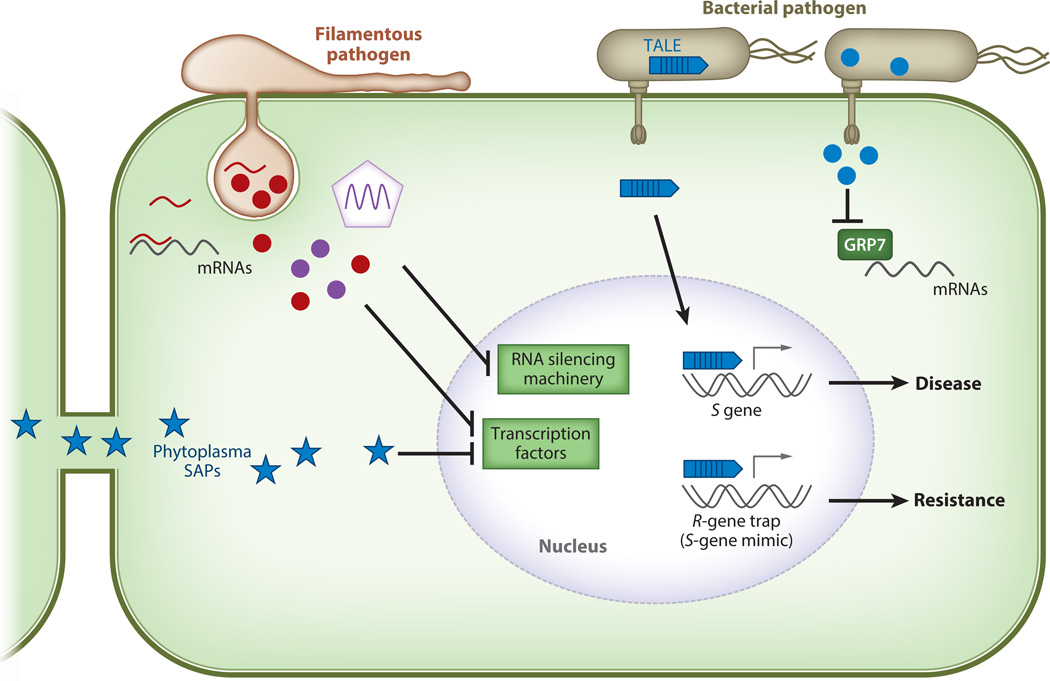
Figure 3.
Pathogen effectors manipulate host gene transcription. Bacterial effectors are depicted in blue, effectors from filamentous pathogens are depicted in red, and viral effectors are depicted in purple. Transcriptional activator-like effectors (TALEs) from Xanthomonas and Ralstonia bacterial pathogens are secreted through the type III secretion system (T3SS) into host cells. TALEs bind to promoter regions of target genes to activate transcription of susceptibility (S) genes and promote virulence. However, some plant genotypes possess resistance (R) genes with TALE-binding promoter regions and act as decoys to activate immune responses. The bacterial pathogen Pseudomonas syringae delivers the T3SS effector HopU1 (blue circle) that targets the RNA-binding protein GRP7. HopU1 interferes with GRP7’s ability to bind messenger RNAs, including immunity-related RNAs. Phytoplasmas are insect-transmitted, phloem-limited bacterial pathogens that secrete SAP effectors (blue star). SAP effectors are able to move throughout the plant via plasmodesmata, and some, such as SAP11 and SAP54, manipulate host transcription factors to induce plant developmental changes favoring insect colonization. Filamentous pathogens (oomycetes and fungi) as well as viruses deliver effectors to suppress plant RNA silencing and thus interfere with plant transcription factors and promote virulence. Botrytis cinerea is able to deliver fungal small RNAs into the host cells that suppress gene expression (red line).