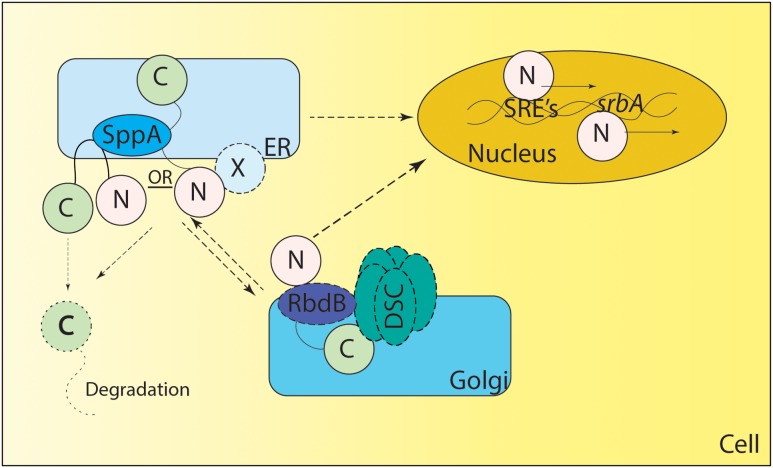
FIGURE 2.
Proposed Model of SREBP regulation in A. fumigatus. SrbA (full length SrbA protein is represented as N terminus and C terminus joined by a transmembrane region) is an ER resident protein, however, the membrane topology of SrbA is unknown. Unlike Schizosaccharomyces pombe, SCAP has not been identified in A. fumigatus, and it is not clear if an unidentified ER resident protein “X” forms a complex with SrbA to regulate SrbA activation. Golgi resident Dsc proteins (DscA-E collectively known as DSC complex) are indispensable for SrbA cleavage and activation, however, it is unclear if there is anterograde and/or retrograde movement of SrbA to the Golgi or Dsc complex movement to the ER for SrbA cleavage. The rhomboid protease RbdB is indispensable for A. fumigatus cleavage and in S. pombe Rbd2 interacts with the UBX domain of Dsc5 (DscE homolog of A. fumigatus) via Cdc48. However, the nature of this interaction needs validation in A. fumigatus. The Signal Peptide Peptidase (SppA) is an ER resident aspartyl protease involved in regulated intramembrane proteolysis and is indispensable for SrbA cleavage, however, it is not clear if SppA cleavage is preceded or followed by action of the Dsc complex and/or RbdB mediated cleavage. Once cleaved, the C terminus of the protein is potentially degraded and the N terminus translocates to the nucleus where it binds to SRE elements in the promoter region of genes involved in the hypoxia response. SrbA also positively regulates its own mRNA levels by binding the SRE element in the promoter region of srbA. Solid lines depict experimentally validated results, whereas dotted lines indicate predicted but not experimentally tested mechanisms. X – Unknown ER resident protein or unidentified SCAP (like) homolog.