Abstract
Genes encoding subunits of SWI/SNF chromatin remodeling complexes are collectively mutated in ~20% of all human cancers1–2. Although ARID1A is the most frequent target of mutations, the mechanism by which its inactivation promotes tumorigenesis is unclear. Here, we demonstrate that Arid1a functions as a tumor suppressor in the mouse colon, but not the small intestine, and that invasive ARID1A-deficient adenocarcinomas resemble human colorectal cancer (CRC). These tumors lack deregulation of APC/β-catenin, crucial gatekeepers in common forms of intestinal cancer. ARID1A normally targets SWI/SNF complexes to enhancers, where they function in coordination with transcription factors (TFs) to facilitate gene activation. ARID1B preserves SWI/SNF function in ARID1A-deficient cells, but defects in SWI/SNF targeting and control of enhancer activity cause extensive dysregulation of gene expression. These findings represent an advance in colon cancer modeling and implicate enhancer-mediated gene regulation as a principal tumor suppressor function of ARID1A.
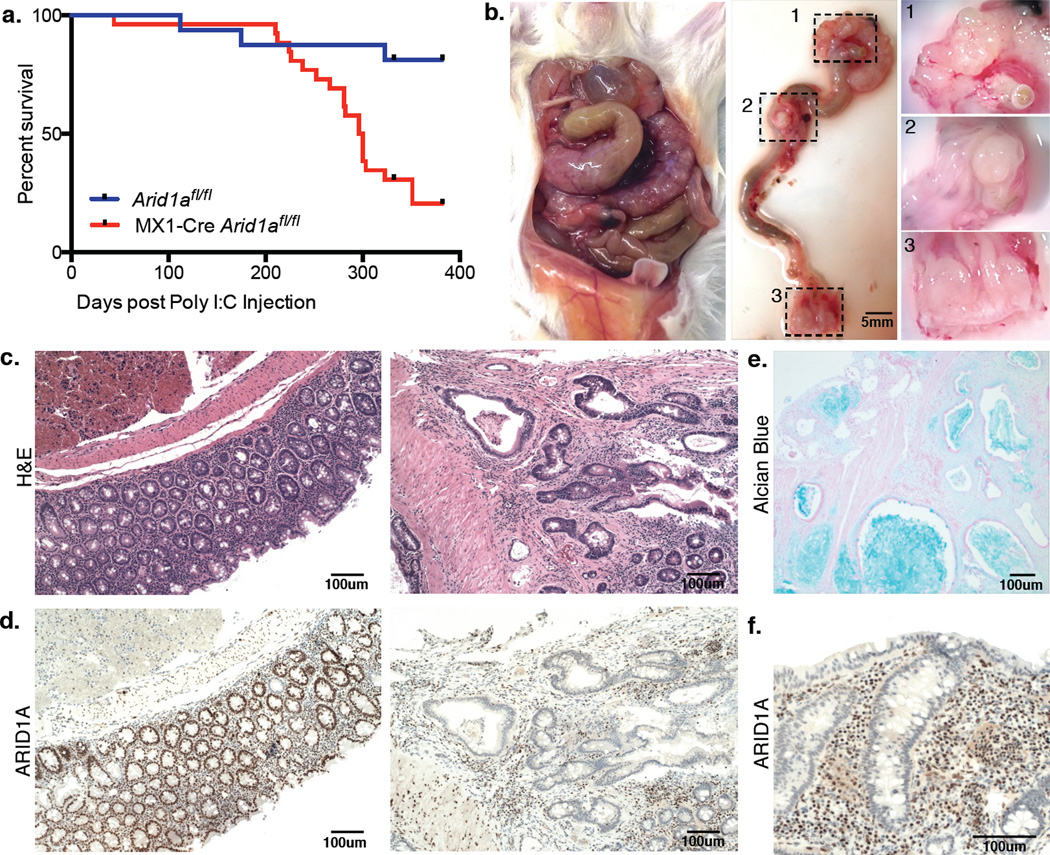
As inactivating ARID1A mutations occur in a broad spectrum of human cancers2, we utilized a mouse model to identify tissues where ARID1A might function as a tumor suppressor in vivo. We generated MX1-Cre Arid1afl/fl mice to achieve sporadic, interferon-responsive inactivation of both Arid1a alleles across many tissues3 (Supplementary Fig. 1a–b). Mice were initially healthy following induction of Cre activity with synthetic interferon (poly I:C), but required euthanasia at a median of 296 days owing to emaciation and rectal prolapse (Fig. 1a, Supplementary Fig. 1c). We identified nodular and polypoid tumors in the colons of these mice, often at multiple non-contiguous sites, but never in the small intestine (Fig. 1b). Tumor histology was consistent with invasive colon adenocarcinoma (Fig. 1c), a malignant neoplasm derived from glandular colonic epithelium4. ARID1A expression was lost in <10% of normal colon epithelial cells, but in all tumor cells (Fig. 1d). Villin-CreER-T2 Arid1afl/fl mice, where Arid1a inactivation was restricted to intestinal epithelial cells5, also developed invasive ARID1A-deficient adenocarcinoma (Supplementary Fig. 2a–d); tumor formation thus reflects epithelial cell-intrinsic ARID1A deficiency. Tumors in Arid1afl/fl mice were marked by prominent mucinous differentiation and presence of tumor-infiltrating lymphocytes (Fig. 1e–f, Supplementary Fig. 2e–g), features associated particularly with human CRCs that show microsatellite-instability (MSI)6. As ARID1A is the third most significantly mutated gene in human CRC, with highest frequency (~39%) in cancers of the MSI type7–8, these findings are highly relevant to human disease.
Figure 1. ARID1A loss drives invasive colon adenocarcinoma in mice.
(a) Survival of MX1-Cre Arid1afl/fl mice (n=26) and control Arid1afl/fl mice (n=16) following injection of Poly I:C;
(b) MX1-Cre Arid1afl/fl mouse with tumors in the (1) cecum, (2) mid-colon, and (3) rectum;
(c) H&E staining on normal colon epithelium (left) and tumor (right) tissue sections from MX1-Cre Arid1afl/fl mouse;
(d) ARID1A immunohistochemistry (IHC) on above tissue sections;
(e) Alcian blue staining on tumor section from MX1-Cre Arid1afl/fl mouse;
(f) ARID1A IHC on tumor section from MX1-Cre Arid1afl/fl mouse showing lymphocytic infiltrate.
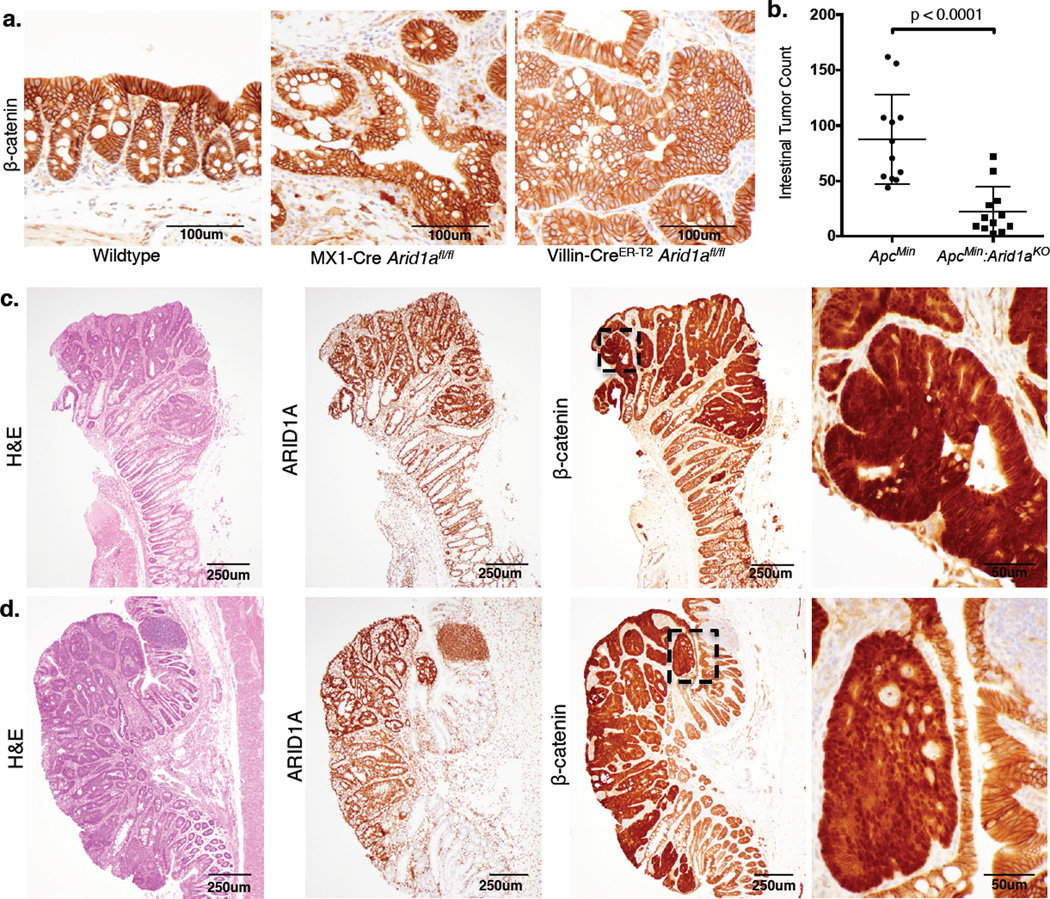
To identify potential cooperating events in tumorigenesis driven by ARID1A deficiency, we obtained whole-exome sequences of DNA isolated from tumor and matched normal tissue from three MX1-Cre Arid1afl/fl mice. Variant analysis of the exome data identified few non-synonymous mutations, none of which were in genes recurrently mutated in human CRC7 (Supplementary Fig. 3, Supplementary Table 1). APC inactivation is an initiating event in human CRC, which drives adenoma formation by allowing β-catenin to translocate to the nucleus and trigger constitutive transcription of Wnt target genes9–10. In Arid1afl/fl tumors, β-catenin localized exclusively at the plasma membrane, indicating intact APC function (Fig. 2a). To characterize further the relationship between ARID1A and APC inactivation in colon tumorigenesis, we obtained mice carrying the germline ApcMin mutation11 and generated ApcMin:Arid1aKO (ApcMin: Villin-CreER-T2 Arid1afl/fl) mice (Supplementary Fig. 4a–b). Whereas ApcMin mice developed the expected number of adenomas throughout the intestines, ApcMin:Arid1aKO mice showed significantly fewer intestinal tumors (Fig. 2b, Supplementary Fig. 4c). The few tumors that did arise in ApcMin:Arid1aKO mice were non-invasive adenomas that were histologically similar to those in ApcMin mice; they retained ARID1A expression and showed nuclear β-catenin localization (Fig. 2c–d, Supplementary Fig. 4d–e). Thus, ARID1A loss drives invasive colon cancer via a mechanism independent of APC inactivation; further, ARID1A is required to facilitate tumorigenesis driven by inactivation of APC.
Figure 2. ARID1A loss drives colon tumorigenesis independent of APC inactivation.
(a) β-catenin IHC in wildtype mouse colon and tumor tissue sections from MX1-Cre Arid1afl/fl and Villin-CreER-T2 Arid1afl/fl mice;
(b) Total tumor count in the small intestine and colon of ApcMin (n=12) and ApcMin:Arid1aKO mice (n=12), p-value shown from unpaired two-tailed T-test;
(c) Colon adenoma in ApcMin mouse with H&E staining, ARID1A IHC, and β-catenin IHC (β-catenin IHC magnification shown for marked tumor region);
(d) Same as (c) for colon adenoma in ApcMin:Arid1aKO mouse.
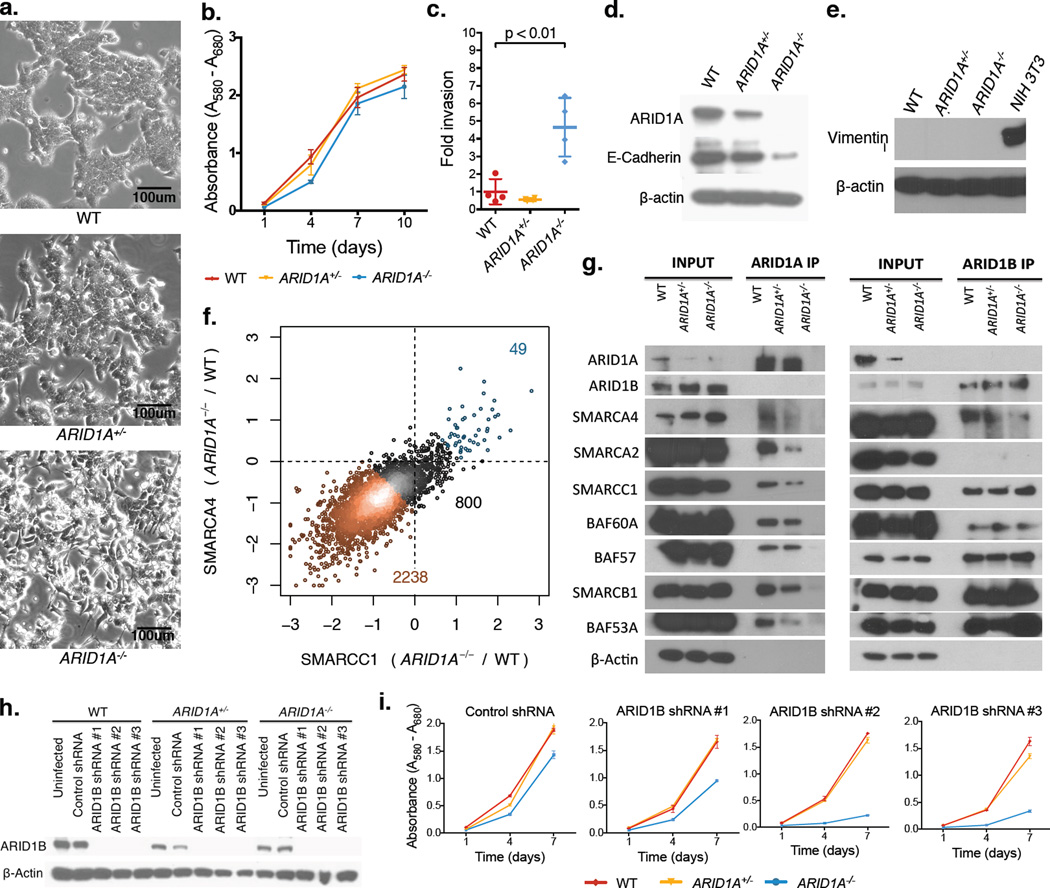
To investigate the tumor suppressor function of ARID1A, we utilized the HCT116 MSI+ human CRC cell line and isogenic lines with mono- (ARID1A+/−) or bi- (Arid1a−/−) allelic deletion of ARID1A. Parental ARID1A-wildtype (WT) cells grew in clustered colonies with tight cell-cell adhesion, while Arid1a−/− cells spread across the culture dish with elongated, spindle-shaped morphologies and frequent filopodia (Fig. 3a). Arid1a−/− cells proliferated normally (Fig. 3b), but showed increased invasiveness (Fig. 3c) and reduced expression of the cell adhesion protein E-Cadherin (Fig. 3d); we did not identify a molecular signature associated with epithelial-mesenchymal transition (EMT, Fig. 3e, Supplementary Fig. 5). Notably, ARID1A depletion causes similar defects in cells derived from human gastric and hepatocellular carcinoma, which also frequently carry inactivating ARID1A mutations12–13.
Figure 3. ARID1A loss causes defects in SWI/SNF targeting to chromatin.
a) Live cell morphology of HCT116 ARID1A WT, ARID1A+/−, and Arid1a−/− cells in culture;
b) Proliferation measured by MTT assay, error bars show standard deviation of 3 measurements (technical replicates);
c) Invasion measured by Matrigel-chamber based assay, error bars show standard deviation of 4 measurements (technical replicates);
d) Protein levels of ARID1A, E-Cadherin, and β-actin;
e) Protein levels of Vimentin and β-actin (Note: NIH 3T3 fibroblast cells included for positive control);
f) Fold change (log2) in SMARCA4 and SMARCC1 ChIP-Seq signals at SWI/SNF binding sites in Arid1a−/− cells relative to WT;
g) Immunoprecipitation of SWI/SNF complexes using antibodies targeting ARID1A and ARID1B;
h) Protein levels of ARID1B and β-actin following ARID1B knockdown with 3 independent shRNAs;
i) Proliferation following shRNA-mediated ARID1B knockdown measured by MTT assay; error bars show standard deviation of 3 measurements (technical replicates).
ARID1A is mutually exclusive in SWI/SNF complexes with ARID1B; both proteins contain DNA binding activity and are implicated in targeting SWI/SNF complexes to chromatin14. To examine consequences of ARID1A loss on SWI/SNF targeting, we performed ChIP-Seq for two core subunits – SMARCA4 (BRG1) and SMARCC1 (BAF155) – in HCT116 WT and Arid1a−/− cells. SMARCA4 and SMARCC1 binding were highly correlated (Supplementary Fig. 6) and we considered sites enriched for both subunits as SWI/SNF binding sites (See Methods, Supplementary Table 2). A large majority of SWI/SNF binding sites (79.2%) showed loss of SMARCA4 and SMARCC1 in Arid1a−/− cells (Fig. 3f). Independent ChIP-qPCR experiments showed that ARID1A was present at these sites in WT cells, consistent with a direct role for this subunit in SWI/SNF targeting (Supplementary Fig. 7, Supplementary Table 3). Few sites gained SMARCA4 and SMARCC1 in Arid1a−/− cells (1.5%, Fig. 3f); these sites also gained ARID1B (Supplementary Fig. 7, Supplementary Table 3). Immunoprecipitation confirmed that ARID1B-containing SWI/SNF complexes were intact in Arid1a−/− cells (Fig. 3g); proliferation of these cells was impaired by ARID1B knockdown (Fig. 3h–i). ARID1A deficiency thus alters SWI/SNF targeting to genomic sites while residual SWI/SNF activity is preserved through ARID1B. Importantly, ARID1B is a synthetic lethal vulnerability in ARID1A-mutant human cancer15–16; cells in Arid1afl/fl tumors also retain expression of ARID1B (Supplementary Fig. 8).
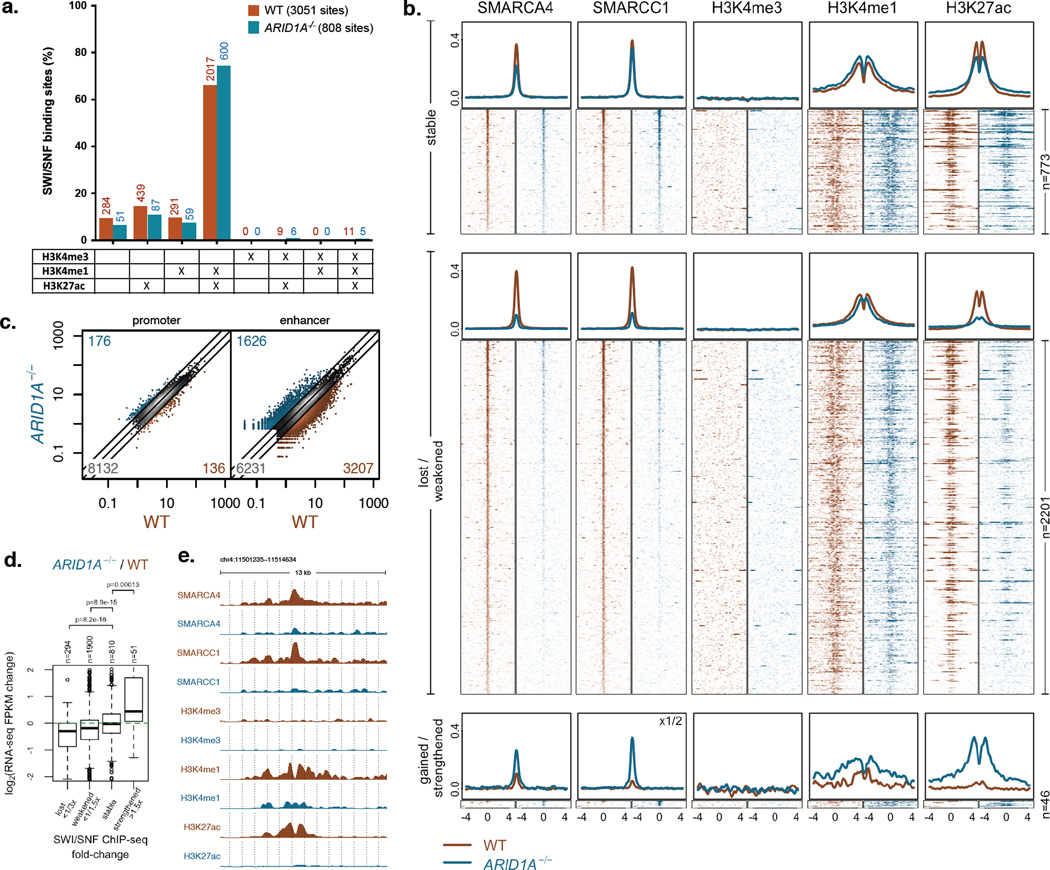
To study consequences of altered SWI/SNF targeting in Arid1a−/− cells, we characterized histone modifications associated with active cis-regulatory elements17 – H3K4me3, H3K4me1, and H3K27ac – at SWI/SNF binding sites in HCT116 WT and Arid1a−/− cells. In both conditions, SWI/SNF binding was highly enriched at active enhancers marked with H3K4me1 and H3K27ac (Fig. 4a); in contrast, SWI/SNF binding was limited at active promoters, defined as transcription start site (TSS)-overlapping regions enriched for H3K4me3 (See Methods, Supplementary Fig. 9). Of note, in TSS-distal regions, H3K27ac was diminished in Arid1a−/− cells at sites that lost SWI/SNF binding and was dramatically increased at the few sites where SWI/SNF binding was gained (Fig. 4b). Genome-wide examination revealed widespread changes in H3K27ac at enhancers, while promoters showed little change (Fig. 4c). Because H3K27ac distinguishes active from poised/inactive enhancers17,18, we asked whether altered SWI/SNF targeting in Arid1a−/− cells affected enhancer activity. Indeed, changes in SWI/SNF binding at TSS-distal sites correlated with changes in mRNA levels of nearest genes as quantified by RNA-Seq (Fig. 4d, Supplementary Table 4). Partial ARID1A deficiency also affected enhancer activity, with ARID1A+/− cells showing intermediate changes in H3K27ac at enhancers and in transcription of the nearest genes (Supplementary Fig. 10). SWI/SNF complexes are capable of mobilizing nucleosomes19,20, and indeed, ARID1A loss changed not only the level of histone modifications at enhancers, but also their locations (Fig. 4b, e). Together, these findings indicate that ARID1A deficiency specifically impairs enhancer configuration and activity, with marked consequences on gene expression.
Figure 4. SWI/SNF complexes are targeted to enhancers and contribute to their activity.
a) Distribution of SWI/SNF binding sites in HCT116 WT and Arid1a−/− cells relative to histone modifications (Note: difference between WT and Arid1a−/− cells is not significant in a paired t-test);
b) ChIP-seq profiles of SMARCA4, SMARCC1, H3K4me3, H3K4me1, and H3K27ac in WT and ARID1A−/− cells around all TSS-distal SWI/SNF binding sites (Notes: labels on the right of the figure indicate number of sites in each category; labels on top right corners indicate any alterations made in scaling of Y-axis);
c) H3K27ac levels in WT and ARID1A−/− cells at TSS-proximal (promoter) and TSS-distal (enhancer) enrichment regions (Note: numbers in the three corners denote numbers of activated (>2×), inactivated (<1/2×), and stable sites);
d) Fold changes (log2) of gene expression between WT and Arid1a−/− cells for genes nearest to TSS-distal SWI/SNF binding sites split based on Arid1a−/− / WT ChIP-Seq signal;
e) ChIP-seq tracks of SMARCA4, SMARCC1, H3K4me3, H3K4me1, and H3K27ac in WT and ARID1A−/− cells.
To determine if control of enhancer activity by SWI/SNF complexes is coordinated with TFs, we analyzed sequence motifs at enhancers sensitive to ARID1A loss. The CTCF motif was relatively depleted in regions of H3K27ac loss, implying CTCF-bound insulator regions may resist modulation, while the AP1 (JUND/FOSL1) motif was most enriched (Fig. 5a, Supplementary Table 5). ChIP-Seq profiles for HCT116 cells, including those generated by ENCODE21 (Encyclopedia of DNA Elements, Supplementary Table 6), revealed further that H3K27ac loss was most strongly associated with sites bound in WT cells by SWI/SNF complexes and/or TFs including AP1, CEBPB, and TEAD4 (Fig. 5b). Among factors assessed in HCT116 cells, binding of these TFs correlated most strongly with SWI/SNF occupancy (Supplementary Fig. 6b) and was higher at enhancers that lost activity than at enhancers that were unaffected by ARID1A deficiency (Fig. 5c). Gene Ontology analysis of nearest genes implicated enhancers that lost activity as regulators of cell adhesion, development, differentiation, and morphogenesis (Supplementary Fig. 11a, Supplementary Table 5). Activity of super-enhancers, key regulators of genes that confer cell identity22, was also affected (Supplementary Fig. 12, Supplementary Table 5). These results establish a broad role for ARID1A in regulating active enhancers in HCT116 cells, converging on dominant TFs that provide a large fraction of tissue-specific gene regulation.
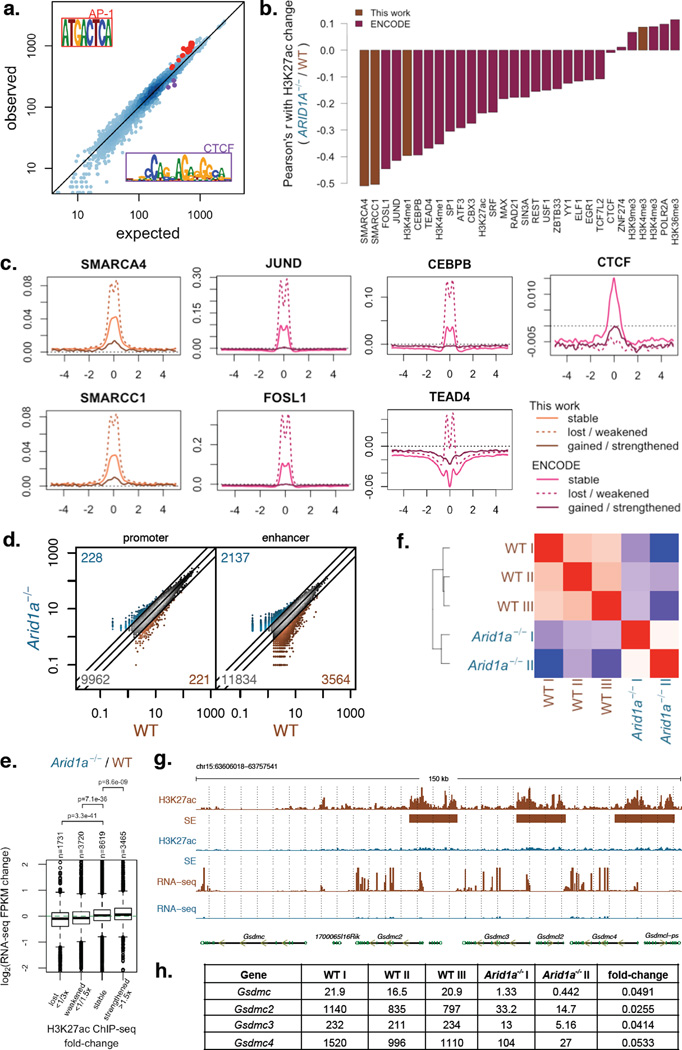
Figure 5. ARID1A loss impairs enhancer-mediated gene regulation in the colonic epithelium.
a) Observed vs. expected TF motif instances at TSS-distal H3K27ac regions (enhancers) with reduced H3K27ac signal based on enrichment regions with stable H3K27ac signal. Motifs highly similar to AP1 and CTCF motifs are highlighted;
b) Correlation between H3K27ac signal change (Arid1a−/− / WT) and WT ChIP-Seq signal levels of different factors profiled in this work and by the ENCODE Project.
c) ChIP-Seq profiles for SMARCA4, SMARCC1, JUND, FOSL1, and CTCF in WT HCT116 cells centered around TSS-distal H3K27ac regions (enhancers) that remain stable, show lost/weakened H3K27ac, or show gained/strengthened H3K27ac in Arid1a−/− cells relative to WT;
d) H3K27ac levels at TSS-proximal (promoter) and TSS-distal (enhancer) enrichment regions for colon epithelium from wildtype (WT) and Villin-CreER-T2 Arid1afl/fl (Arid1a−/−) mice (Note: numbers in the three corners denote numbers of activated (>2×), inactivated (<1/2×) and stable sites);
e) Fold changes (log2) of gene expression between WT and Arid1a−/− mouse colon epithelium for genes nearest to TSS-distal H3K27ac regions (enhancers) split based on Arid1a−/− / WT ChIP-Seq signal;
f) Pearson's correlation among RNA-Seq samples based on FPKM values for mouse colon epithelium dissociated from individual wildtype mice (WT, n=3) and Villin-CreER-T2 Arid1afl/fl mice (Arid1a−/−, n=2);
G) H3K7ac ChIP-Seq tracks, super-enhancer (SE) calls, and RNA-Seq tracks at Gsdmc locus in WT and Arid1a−/− mouse colon epithelium.
H) RNA-Seq FKPM values for individual WT and Arid1a−/− mice for Gsdmc, Gsdmc2, Gsdmc3, and Gsdmc4.
To determine if ARID1A loss impairs enhancer activity in vivo, we examined H3K27ac and gene expression in the colonic epithelium of wildtype and Villin-CreER-T2 Arid1afl/fl mice. Again, whereas promoter activation states were changed little (Fig. 5d), the effects on H3K27ac at enhancers were significant and were correlated with changes in mRNA levels of the nearest genes (Fig. 5e). Developmental enhancers were identified as most sensitive to ARID1A loss, showing many Gene Ontology terms shared with HCT116 cells (Supplementary Fig. 11b, Supplementary Table 7). More than 1,000 genes were dysregulated >2-fold in Arid1a−/− colonic epithelium (FDR<0.05), including several genes known to be associated with cancer (Fig. 5f, Supplementary Table 8). For example, the Gasdermins, established tumor suppressors in gastric epithelium23–24, were markedly downregulated (Fig. 5g–h). Although it is difficult to attribute tumorigenesis to discrete target genes, our findings implicate broad control of enhancer activity as the SWI/SNF function crucially impaired in ARID1A-deficient cancers.
Despite the characterization of APC as a gatekeeper in human CRC9–10, normal patterns of β-catenin staining have long been observed in tumors with MSI25. Indeed, recent molecular classification reveals that APC mutations occur with relatively low frequency in the subtype of human CRC (consensus molecular subtype, CMS 1: MSI, Immune) enriched for mutations in ARID1A26. Importantly, not only are tumors in Arid1afl/fl mice histologically distinct from those that develop with APC inactivation, but also they reflect with greater accuracy features of human CRC – aggressive local tissue invasion, long latency, exclusive origin in the colon – that are absent in current models11. Arid1afl/fl mice thus establish a novel pathway to colon tumorigenesis and provide a sound model for further investigation of cell-intrinsic and extrinsic factors that might contribute to colon tumor formation, progression to invasive cancer, and response to therapy.
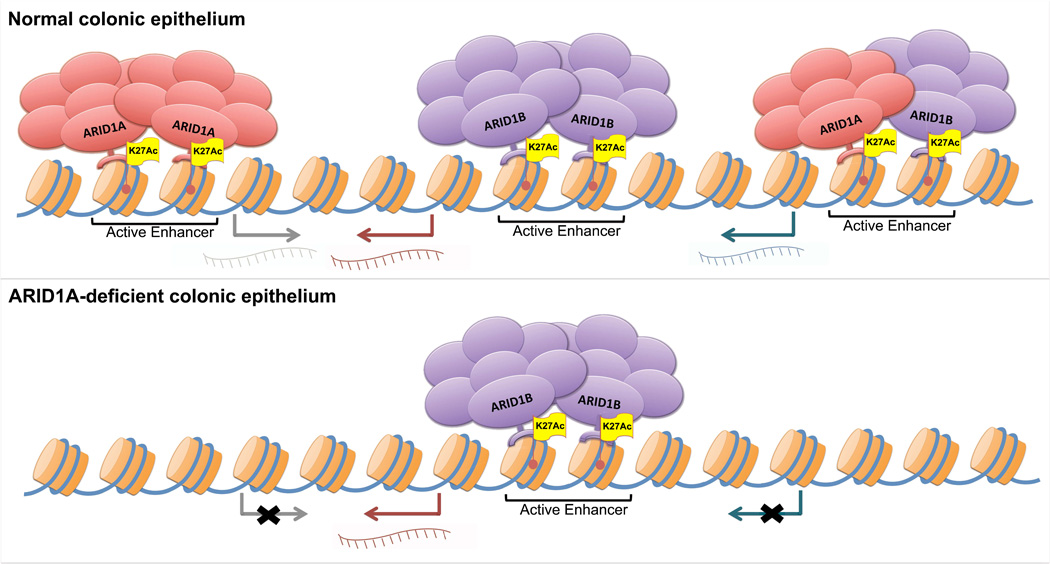
While SWI/SNF function is described at both promoters and enhancers27–29, our findings implicate enhancers as principal sites at which SWI/SNF complexes function to regulate gene activation. When ARID1A is absent, SWI/SNF is lost from thousands of enhancers that subsequently lose activity – showing reduced H3K27ac and expression of nearest genes – while residual SWI/SNF complexes containing ARID1B bind enhancers that remain active (Fig. 6). Enhancers are major determinants of cell type specificity in gene expression28; ARID1A loss impairs SWI/SNF control of enhancers bound by dominant TFs that activate gene expression programs critical for development/differentiation. As SWI/SNF complexes regulate gene expression across lineages and developmental states31,32, defective SWI/SNF control of enhancer activity may underlie not only the oncogenic drive of ARID1A-mutant CRC, but also of other human malignancies driven by alterations in ARID1A and other SWI/SNF subunits.
Figure 6. Defective SWI/SNF targeting and control of enhancer activity in the ARID1A-deficient colonic epithelium (Model)!
Online Methods
Mouse experiments
All experiments were performed with strict adherence to our IACUC-approved Animal Experimentation Protocol #12-017 and guidelines of the Dana-Farber Animal Resource Facility (ARF).
MX1-Cre Arid1afl/fl mice
MX1-Cre mice purchased from Jackson Labs (Stock number 003556) were bred to Arid1afl/fl mice obtained from Dr. Zhong Wang33. 6–8 week old MX1-Cre Arid1afl/fl and littermate control Arid1afl/fl mice were administered pI-pC (polyinosinic-polycytidylic acid, Invivogen tlrl-pic) via intraperitoneal injection at 25ug/g every other day for 7 days. Excision of Arid1a was evaluated in mice 1-week post injection by PCR of DNA harvested from mouse tissues using primers flanking the floxed exon (5'-GTAATGGGAAAGCGACTACTGGAG-3' and 5'-TGTTCATTTTTGTGGCGGGAG-3', See Supplementary Fig. 1a–b). Mice were monitored for health and euthanized upon instruction by the ARF veterinarian staff. Whole mouse necroscopies were conducted on the first cohort of mice at the Dana-Farber/Harvard Cancer Center Rodent Pathology core. Sample size for survival analysis was calculated using estimated effect size from this cohort of mice; no animals were excluded. Formalin-fixed intestines were processed for histology and immunohistochemistry at the DFCI Specialized Histopathology Core.
Villin-CreER-T2 Arid1afl/fl mice
5–7 week old Villin-CreER-T2 Arid1afl/fl mice and littermate control Arid1afl/fl mice were administered 1mg Tamoxifen (T5648 SIGMA) dissolved in sunflower oil (S5007 SIGMA) via intraperitoneal injection for 5 consecutive days. Three Villin-CreER-T2 Arid1afl/fl mice and three control Arid1afl/fl mice were euthanized 8 weeks post-injection for dissociation of colon epithelial cells34. Specifically, Mouse colons were dissected, flushed, and splayed. Colon pieces were rinsed in PBS, incubated in 5mM EDTA, and shaken for 2 minutes to release dissociated epithelial cells. Additional mice were aged for tumor analysis. Intestines were formalin-fixed and processed for histology and immunohistochemistry at the DFCI Specialized Histopathology Core.
ApcMin and ApcMin:Arid1aKO mice
ApcMin mice purchased from Jackson labs (C57BL/6J-Apc-min/J; stock number 002020) were bred to Villin-CreER-T2 Arid1afl/fl mice to generate ApcMin: Villin-CreER-T2 Arid1afl/fl (ApcMin:Arid1aKO) mice and littermate control ApcMin Arid1afl/fl (ApcMin) mice. Mice aged 5–6 weeks were injected intraperitoneally with 1mg Tamoxifen (T5648 SIGMA) dissolved in sunflower oil (S5007 SIGMA) for 5 consecutive days. Age-matched ApcMin and ApcMin:Arid1aKO mice were euthanized at 5–6 months. Blinded counts of intestinal tumors were obtained under a dissecting microscope. A two-tailed t-test was used to evaluate statistical significance, F-test to compare variances. Intestines were formalin-fixed and processed for histology and immunohistochemistry at the DFCI Specialized Histopathology Core.
Whole exome sequencing
DNA from flash-frozen matched tumor and tail tissue from MX1-Cre Arid1afl/fl mice was purified with the DNEasy Blood & Tissue Kit (Qiagen 69504) following histological confirmation of invasive adenocarcinoma. Samples were further processed and analyzed at the Dana-Farber Center for Cancer Computational Biology (CCCB). Target enrichment was performed using the SureSelectXT Mouse All Exon (Agilent Technologies) bait library. Samples were sequenced on the Next-Seq500 (Illumina) system using the PE-150 flowcell. Following sequencing and demultiplexing, sequencing reads were trimmed such that the lowest quartile of the phred-scaled Q-score was greater than 28; typical read lengths were approximately 140bp. The paired reads were aligned to the Ensembl GRCm38.75 genome using BWA-mem35–36 using default parameters. Following initial alignment, reads were further processed using GATK best-practices for WES data, including marking of duplicates, de novo realignment near putative indels, and base-quality score recalibration.
Variant calling was performed with VarScan2 software37 (v2.4.1), due to the paired (tumor/normal) design of the experiment. Somatic, germline, and loss-of-heterozygosity events are reported. Default parameters were used, requiring a minimum coverage depth of 8 reads for variant calls. In addition to the set of VarScan2 calls, MuTect software38 was used to generate a second set of somatic point mutations.
VarScan2 was also used for generation of putative copy-number variations between the matched samples. Following the recommendation of the documentation, CNV calls were filtered and finely smoothed/segmented using Bioconductor's DNACopy package, which implements the CBS algorithm. Additionally, a second set of CNV calls was generated using CONTRA software39 with default parameters and specifying the targeted regions from the exome capture process.
Cell line experiments
Cell culture
The HCT116 cell line and derivative ARID1A+/− and Arid1a−/− isogenic cell lines were purchased from Horizon Discovery (HD 104-031 and HD 104-049) and cultured in RPMI 1640 supplemented with 10% FBS as per instructions. These cell lines were negative for mycoplasma and all other infectious agents evaluated under the Mouse/Rat Comprehensive CLEAR Panel (Charles River Research Animal Diagnostic Services).
Knockdown experiments
Lentiviral shRNAs on the PLKO.1 vector targeting ARID1B (clone #1: TRCN0000107361, clone #2: TRCN0000107363, clone #3: TRCN0000107364) were used to infect into HCT116 WT, ARID1A+/− and Arid1a−/−cells. Infected cells were selected in Puromycin for 72 hours and then plated in triplicate onto an MTT assay (Cell Proliferation Kit, Roche # 1465007001) with 5000 cells/well.
Western blotting and co-immunoprecipitation
Whole cell extracts of isogenic HCT116 cell lines were used in Western blots for E-Cadherin (Cell Signaling Technology: 24E10), ARID1A (Cell Signaling Technology: 12354), ARID1B (ABCAM ab57461) and ACTIN (Cell Signaling Technology: 5125). Nuclear extracts for co-immunoprecipitation were prepared using the NE-PER Nuclear and Cytoplasmic Extraction Kit (Thermo Scientific #78835). Nuclear extracts were diluted with RIPA buffer (Life Technologies 89900) to a final concentration of 1 mg/ml (with protease inhibitor cocktails, Roche). Each IP was incubated with SMARCC1/BAF155 antibody (Santa Cruz: sc9746), ARID1A antibody (Millipore PSG3), or ARID1B antibody (Santa Cruz 32762) overnight at 4°C. Protein G Dynabeads (Life Technologies 10009D) were added and incubated at 4°C for 3 h. Beads were then washed three times with RIPA buffer and resuspended in reducing SDS gel loading buffer. Antibodies to the following proteins were used in the immunoblots: ARID1A (Cell Signaling Technology: 12354); ARID1B (Abcam: ab54761); SMARCA4/BRG1 (Santa Cruz: sc17796); BRM (Cell Signaling Technology: 11966); SMARCC2/BAF170 (Bethyl Laboratories: A301-039A); SMARCD1/BAF60A (Bethyl Laboratories: A301-595A); SMARCE1/ BAF57 (Bethyl Laboratories: A300-810A); SMARCB1/SNF5 (Bethyl Laboratories: A301-087A); ACTL6A/BAF53A (Bethyl Laboratories: A301-391A); ACTIN (Cell Signaling Technology: 5125, 1:3,000).
Invasion assay
25,000 serum-starved cells were added to the top of each BioCoat Matrigel Invasion Chamber (BD Biosciences #354480) with chemoattractant (RPMI, 10% FBS) at the bottom of wells. Chambers were incubated for 22 hours at 37°C, 5% CO2. Non-invading cells were removed from upper surface of the membrane by scrubbing with cotton tipped swabs. Blind counts of invaded cells were obtained following Crystal Violet staining of live cells. This assay was performed with 4 replicates for each cell type. Two-tailed t-tests were used to determine statistical significance, F tests to compare variances.
Sample preparation for ChIP-Seq and ChIP-qPCR
HCT116 isogenic cell lines were dual-crosslinked in 2mM disuccinimidyl glutarate (DSG; Life Technologies #20593) for 30 min then 1% formaldehyde for 10 min, followed by 5 min glycine quenching. Nuclear extracts were generated following 3 washes in PBS. Chromatin was fragmented using Covaris sonication (adaptive focused acoustics; AFA technology). The following antibodies were used for immunoprecipitation of 30ug solubilized chromatin: SMARCC1/BAF155 (Santa Cruz sc9746; ChIP-Seq and ChIP-qPCR), SMARCA4/BRG1 (Abcam ab110641; ChIP-Seq and ChIP-qPCR), H3K27ac (Abcam ab4729; ChIP-Seq and ChIP-qPCR); H3K4me (Abcam ab8895; ChIP-Seq), H3K4me3 (Abcam ab8580; ChIP-Seq); ARID1A (Santa Cruz 32761 and Millipore PSG3; ChIP-qPCR) and ARID1B (Abcam ab57461 and Santa Cruz 32762; ChIP-qPCR). Antibody:chromatin complexes were pulled down with Protein G dynabeads (Life Technologies 10004D), washed, and eluted. Chromatin crosslinks were reversed and samples were treated with Proteinase K and RNAse A. ChIP-DNA was extracted with the Min-Elute PCR purification kit (Qiagen) and quantified with Quant-iT™ PicoGreen dsDNA Assay Kit (Life Technologies). 10ng of purified ChIP-DNA was used to prepare sequencing libraries for the Illumina Hi-Seq Genome Analyzer. ChIP-qPCR was performed on the ViiA7 Real-Time PCR System (Life Technologies) using SYBR Select Master Mix (Life Technologies) using 1ul of purified ChIP-DNA in duplicate in 384-well format. A minimum of two independent replicate experiments were performed for each factor analyzed by ChIP-qPCR.
Dissociated colon epithelial cells pooled from 3 Villin-CreER-T2 Arid1afl/fl mice and from 3 control Arid1afl/fl mice were fixed in 2% formaldehyde, lysed, and pulsed at 15% amplitude on a tip-sonicator. Sonicated chromatin was immunoprecipitated using antibodies for H3K27ac (Abcam ab4729) and H3K4me3 (Abcam ab8580). Antibody:chromatin complexes were pulled down with Protein G dynabeads (Life Technologies 10004D), washed, and eluted. Chromatin crosslinks were reversed and samples were treated with Proteinase K and RNAse A. ChIP-DNA was extracted with the Min-Elute PCR purification kit (Qiagen) and quantified with Quant-iT™ PicoGreen dsDNA Assay Kit (Life Technologies). 10ng of purified ChIP-DNA was used to prepare sequencing libraries for the Illumina Hi-Seq Genome Analyzer.
Sample preparation for RNA-Seq
Trizol Reagent (Life Technologies 15596-026) was used to isolate RNA from harvested HCT116 isogenic cell lines and dissociated mouse colon epithelial cells. RNA was further purified with the RNeasy Mini Kit (Qiagen) and Turbo DNA-Free Kit (Ambion; murine cells only). Sequencing libraries were generated using Tru-Seq Technology (Illumina) for the Illumina Hi-seq Genome Analyzer.
ChIP-qPCR Processing
ChIP-qPCR signals were normalized using the percent-input method: [100*2^(Adjusted input - Ct (IP)]. For each independent replicate experiment, statistical significance in WT v. Arid1a−/− ChIP-qPCR signal was determined using the Holm-Sidak method, with alpha=5.000% (GraphPad Prism version 6 for Mac OS X). Computations assume that all rows (individual ChIP-qPCR sites) are sample from populations with the same scatter (SD).
Log-fold-change for each independent replicate experiment was calculated using the ratio of percent input for ARID1A−/− over WT (averaged over technical replicates). Averages of ARID1A−/−/WT log-fold-change values were calculated for each factor from all independent replicate experiments for Supplementary Fig. 7b–c.
ChIP-Seq Processing
Alignment, fragment size estimation, and library complexity
The sequenced reads were aligned to the hg19/mm9 genome assembly using Bowtie40 0.12.6, allowing up to 10 matches (‘-m 10 --best’ options). For HCT116, reads on the 24 assembled chromosomes excluding the ENCODE blacklisted regions were kept for downstream analysis. For the mouse samples, reads on the 19 assembled autosomes excluding a custom 280kb blacklist region were kept for downstream analysis. The custom blacklist regions were selected based on very high signal in input tracks in a parallel study (GSE71509). Peaks of cross-correlation profiles were identified to estimate the typical fragment size for each sample. The typical fragment size for the different samples ranged between 140–180bp. Each read was considered to represent a signal at half typical fragment size from the 5' end. Library complexity was calculated for each sample as the number of unique bp positions mapped on each strand, divided by the total number of mapped reads. For batches of experiments where the typical library complexity was below 90% (all mouse samples, HCT116 H3K4me1/3 and matching input), only one read mapping to each position was kept.
Identification of regions of enrichment (RoE)
Different ChIP-seq regions of enrichment (RoE) were identified using the SPP package41 in R, with the function get.broad.enrichment.clusters and option window.size=500, with matching input samples for each IP experiment, using appropriate z.thr values for each analysis as specified below. The input samples for samples for WT and ARID1A-deficient mice had relatively low sequencing depth (5.7 and 12.0M reads after selecting one read per position). The two samples appeared to show no more systematic variability than expected from the statistical variability due to low sequencing depth. Therefore the two were merged and the combined input was provided to SPP as a control for both WT and ARID1A-deficient H3K27ac ChIP-seq.
Defining active TSSs and H3K4me3 RoEs
Active TSSs in HCT116 were defined as all TSSs in Ensembl release GRCh37.72 that overlapped H3K4me3 RoEs (z>4) in either condition. Active TSSs in mouse colon were defined as all TSSs in Ensembl release NCBIM37.67 that overlapped an H3K27ac (z>4) peak in either condition (H3K4me3 ChIP-seq failed for ARID1A-deficient mouse colon; the enrichment at TSSs was relatively low for the WT colon sample). Additionally, H3K4me3 RoEs in mouse colon were defined as the union of RoEs (z>4) called with the WT colon sample and a WT MEF sample from the accompanying study (GSE71509). These two regions, i.e., "active TSSs" and "H3K4me3 RoEs" were used together to conservatively define TSS-proximal and TSS-distal regions in downstream analyses, as specified below.
Identification and classification of SWI/SNF binding sites
SWI/SNF binding sites were identified in two steps: First, overlapping SMARCA4 and SMARCC1 RoEs with z>4 were called for each condition, WT and ARID1A−/−. Next, the union of the regions for the two conditions was calculated. This approach reduces any bias that may arise in differential RoE calling due to thresholds. The sites that were called in WT, or those where the signal in WT was more than half the signal in ARID1A−/− were considered as SWI/SNF binding sites in WT cells; the complementary selection was performed for SWI/SNF binding sites in Arid1a−/− (used in Fig. 4a). SWI/SNF binding sites overlapping both an active TSS and an H3K4me3 RoE were called TSS-proximal. Those more than 1kb away from an H3K4me3 RoE and more than 2kb away from an active TSS were called TSS-distal. Others were ambiguous and excluded from studies specific to TSS-proximal or distal sites. When we evaluated where SWI/SNF binding falls in the genome, we used H3K27ac and H3K4me1 RoEs that are called inclusively with a z>3 threshold. Changes in TSS-distal SWI/SNF binding upon ARID1A loss were evaluated at each binding site, by dividing the library-size normalized IP signal for SMARCA4 and SMARCC1 in ARID1A−/− by WT. If the geometric mean of change was greater than 1.5 fold, and both factors showed increased signal, the RoE was called as gained/strengthened. In reverse, if the geometric mean of change was less than 2/3 fold, and both factors showed decreased signal, the RoE was called as lost/weakened. Other sites were called as unchanged (See colors in Fig. 3f).
Identification and classification of H3K27ac Roes (promoters and enhancers)
H3K27ac RoEs with z>4 were called to specifically study changes in this mark in HCT116 and colon epithelial cells (Fig. 4c, 5e). Similar to SWI/SNF binding sites, H3K27ac RoEs from different conditions were merged, ones overlapping both an active TSS and an H3K4me3 RoE were defined as promoters; and ones that were more than 1kb away from an H3K4me3 RoE and more than 2kb away from an active TSS were called as enhancers.
Gene ontology analysis for enhancers
To identify enhancers that lost SWI/SNF binding, we used an inclusive definition of SWI/SNF binding as overlapping SMARCA4 and SMARCC1 RoEs with a z>3 threshold. Enhancers with SWI/SNF binding, where the average SMARCA4 and SMARCC1 signal was down more than 1.5 fold were called as SWI/SNF losing enhancers. GO analysis for these SWI/SNF losing enhancers was performed as follows: Gene Ontology databases were downloaded from geneontology.org on 2014/04/29. Each enhancer was associated to the closest active TSS within 100kb. p-values for gene set enrichment for genes associated to SWI/SNF losing enhancers were calculated relative to genes associated with any enhancer using hypergeometric test. q-values were obtained based on Benjamini–Hochberg procedure.
IP efficiency correction for H3K27ac samples
The efficacy of IP pull-down may vary between different ChIP-seq experiments. A number of lines of evidence suggested that the real levels of H3K27ac are unchanged at a large fraction of promoters upon ARID1A deletion: i. we found that a large fraction of promoters show the same amount of fold-change with very small variance; ii. the typical fold-change was independent of SWI/SNF binding at promoters, and was the same as at enhancers with no SWI/SNF binding; iii. in the accompanying MEF study, we observed different fold-changes at promoters, both greater and less than one-fold for replicates of experiments upon Smarcb1 knockout, while we saw consistent decrease of H3K27ac in western blots and at enhancers. Based on these observations, we applied a small multiplicative factor on H3K27ac samples to set the mode of the log-fold-change distribution at promoters to zero while comparing WT and ARID1A-deficient samples. These factors were HCT116: Parental: 0.95, ARID1A−/− : 1.31, ARID1A+/− : 0.82; Mouse colon epithelial cells: WT: 1.08, ARID1A-deficient: 0.92. This normalization procedure does not affect the qualitative observations presented. We refrained from applying a similar normalization for other ChIP-seq sample pairs, since we could not confidently determine a set of regions where they are unaffected upon ARID1A loss.
ChIP-seq visualization
Genomic profiles for visualization were generated using Gaussian smoothing with sigma=100bp after library size normalization (e.g. in Fig. 4e, or output wig files). The SWI/SNF binding site heatmaps (e.g. in Fig. 4b) were centered at the position with highest signal in the smoothed profile obtained by summing the four tracks considered, (WT or Arid1a−/−, SMARCA4 and SMARCC1). The heatmaps show input subtracted values, whereas the browser shots show raw smoothed signal. The average profiles for each class in Fig. 4b (e.g. unchanged) were obtained as 0.1–0.9 trimmed linear mean at each position.
Identification of super-enhancers
Super-enhancers were called with a slightly modified approach from the original method22. H3K27ac RoEs were called as described above. We did not remove TSS-proximal peaks, but stitched all RoEs within 12.5kb. For each stitched RoE, IP and input signal were calculated only in portions that did not intersect H3K4me3 RoEs (as defined above). We found this approach to be better at removing false positives from stitched enhancer peaks which encompassed active TSSs. Each super-enhancer was associated to the closest active TSS, as defined above, within 300kb. If more than one gene is similarly close to a super-enhancer (at most 50kb further than the closest gene), those genes are also listed in the super-enhancer associated gene list. The inclusive z>3 threshold was used to identify SWI/SNF binding sites within super-enhancers in Supplementary Fig. 12c.
Transcription Factor Motif Enrichment
Transcription factor motif maps for hg19 for 4095 motifs (including a redundant set of real transcription factor recognition elements, and shuffled motif control sequences) were downloaded from http://compbio.mit.edu/encode-motifs/42. The position weight matrix (pwm) for each motif was calculated based on the actual sequences of the provided motif locations. The number of motif occurrences was counted for each motif inside lost/weakened enhancers (H3K27ac fold change< 1/1.5) and unchanged enhancers (1/1.5<H3K27ac fold-change<1.5). A lowess curve was calculated to model the ratio of counts for each motif (sensitive/insensitive) as a function of the GC content of the motif pwm. This curve was used to calculate back the null hypothesis expected number of occurrences for each motif in the lost/weakened. Figures 5a shows observed counts vs. expected counts for the 4095 motifs. pwms for two selected motifs (“AP-1_known3_8mer” and “CTCF_known1_8mer”) are displayed on the figure. Motif similarity was assessed based on Pearson correlation values between motif pwms; motifs which are similar to the two selected motifs (r>0.85) are highlighted on the figure.
Correlation of binding of different factors in WT with H3K27ac changes
ChIP-seq signal around the peak point of each H3K27ac RoE was calculated within +/−1.5kb, per million mapped reads; a pseudocount of 1 was added, and the values were logged (L=log2(1+signal)). The change in H3K27ac was calculated using samples generated in this work as L(ARID1A)-L(WT). For each factor from ENCODE project, the L values were averaged over replicates at each H3K27ac RoE. The Pearson’s correlation between each factor and the change in H3K27ac is plotted.
RNA-seq processing
The sequenced reads from each sample were aligned to the human/mouse genome+transcriptome assemblies GRCh37.72/NCBIM37.67 using TopHat43 v2.0.8 with default parameters except turning off novel junction search (‘-G <gtf> --no-novel-juncs’ options). The transcriptome was self-merged to allow processing with cufflinks44 v2.1.1 tool cuffdiff, “cuffcompare -s hg19.fa -CG -r GRCh37.72.gtf GRCh37.72.gtf” (and similarly for mouse). Different conditions were compared using cuffdiff with default parameters and bias correction (‘-G <gtf> -b’ options).
Each SWI/SNF or H3K27ac RoE was associated to the closest active TSS as defined above. The connection between ChIP-seq signal change and RNA-seq change (Figures 4d, 5f) was studied for TSS-distal RoEs. Only RoEs for which the closest active TSS is between 5kb and 100kb are retained. IP signal was quantified as ‘total IP signal in RoE per billion mapped reads + pseudocount of 0.1’. RNA signal was quantified as ‘normalized gene level count value from cuffdiff + 5’. The ratio of IP signal for Arid1a-deficient divided by WT was used to categorize RoEs to four groups: more than 3-fold signal loss, between 1.5 to 3-fold signal loss, less than 1.5-fold change, and more than 1.5-fold signal increase. The ratio of RNA-seq signal for Arid1a-deficient divided by WT signal was plotted for each category.
The heatmaps for RNA-seq results show mean-shifted log expression values: log2(normalized gene level count value from cuffdiff + 5) − log2(average value for the gene across all samples).
Supplementary Material
Whole exome sequencing of tumors from MX1-Cre Arid1afl/fl mice
SWI/SNF binding sites in HCT116 cells
ChIP-qPCR data for HCT116 cells
RNA-Seq data with GSEA for HCT116 cells 14
H3K27ac regions of enrichment with nearest genes and gene ontology (GO), motif, super-enhancer (SE) analysis in HCT116 cells
ENCODE accession codes for HCT116 cells
H3K27ac regions of enrichment with nearest genes and GO, motif, SE analysis in mouse colon epithelium
RNA-Seq data with GSEA for mouse colon epithelium
Acknowledgments
We thank Sylvie Robine for providing Villin-CreER-T2 transgenic mice, S.H. Orkin for guidance, and members of the Roberts and Orkin labs for discussion. This work was supported by US National Institutes of Health grants R01CA172152 (C.W.M.R.) and R01DK081113 (R.A.S), by a Claudia Adams Barr grant (C.W.M.R.), and by an Innovation Award from Alex’s Lemonade Stand (C.W.M.R). R.M. and A.K.S.R. were supported by the US National Institutes of Health predoctoral fellowships (1F31CA199994 and 1F31CA180784). X.W. was supported by the Pathway to Independence Award from the US National Institutes of Health (K99CA197640). The Cure AT/RT Now foundation, the Avalanna Fund, the Garrett B. Smith Foundation, Miles for Mary (C.W.M.R.), and the Lind Family (R.A.S.) provided additional support. We also thank the Dana-Farber/Harvard Cancer Center cores in Rodent Histopathology and Specialized Histopathology, which are supported in part by a NCI Cancer Center Support Grant P30CA06516.
Footnotes
Accession codes
All sequencing data are deposited in the GEO database (http://www.ncbi.nlm.nih.gov/geo/) with accession number GSE71514.
Author Contributions
R.M., B.G.W., R.A.S, and C.W.M.R conceived experiments and study design. Mouse and cell line experiments were performed by R.M., A.K.S.R., X.W., and B.G.W. Histopathological analysis was conducted by A.T.A. Computational and statistical analysis was performed by B.H.A with guidance from P.J.P. All authors contributed to data analysis and interpretation. R.M., R.A.S., and C.W.M.R. wrote the manuscript with input from all authors.
The authors declare no competing financial interests.
References
- 1.Garraway LA, Lander ES. Lessons from the Cancer Genome. Cell. 2013;153(1):17–37. doi: 10.1016/j.cell.2013.03.002. http://doi.org/10.1016/j.cell.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 2.Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J, Crabtree GR. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nature Publishing Group. 2013;45(6):592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kühn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269(5229):1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 4.Hamilton SR, Bosman FT, Boffetta P, et al. Carcinoma of the colon and rectum. In: Bosman FT, Carneiro F, Hruban RH, Theise ND, editors. WHO Classification of Tumours of the Digestive System. Lyon: IARC Press; 2010. pp. 134–146. [Google Scholar]
- 5.Marjou el F, Janssen K-P, Chang BH-J, Li M, Hindie V, Chan L, et al. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis (New York, N.Y. : 2000) 2004;39(3):186–193. doi: 10.1002/gene.20042. http://doi.org/10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- 6.Greenson JK, Huang S-C, Herron C, Moreno V, Bonner JD, Tomsho LP, et al. Pathologic Predictors of Microsatellite Instability in Colorectal Cancer. The American Journal of Surgical Pathology. 2009;33(1):126–133. doi: 10.1097/PAS.0b013e31817ec2b1. http://doi.org/10.1097/PAS.0b013e31817ec2b1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. doi: 10.1038/nature11252. http://doi.org/10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cajuso T, Hänninen UA, Kondelin J, Gylfe AE, Tanskanen T, Katainen R, et al. Exome sequencing reveals frequent inactivating mutations in ARID1A, ARID1B, ARID2 and ARID4A in microsatellite unstable colorectal cancer. International Journal of Cancer. 2014;135(3):611–623. doi: 10.1002/ijc.28705. http://doi.org/10.1002/ijc.28705. [DOI] [PubMed] [Google Scholar]
- 9.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61(5):759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 10.Fearon ER. Molecular Genetics of Colorectal Cancer. Annual Review of Pathology: Mechanisms of Disease. 2011;6(1):479–507. doi: 10.1146/annurev-pathol-011110-130235. [DOI] [PubMed] [Google Scholar]
- 11.McCart AE, Vickaryous NK, Silver A. Apc mice: models, modifiers and mutants. Pathology, Research and Practice. 2008;204(7):479–490. doi: 10.1016/j.prp.2008.03.004. http://doi.org/10.1016/j.prp.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 12.Yan H-B, Wang X-F, Zhang Q, Tang Z-Q, Jiang Y-H, Fan H-Z. Reduced expression of the chromatin remodeling gene ARID1A enhances gastric cancer cell migration and invasion via downregulation of E-cadherin transcription. Carcinogenesis. 2014;35(4):867–876. doi: 10.1093/carcin/bgt398. http://doi.org/10.1093/carcin/bgt398. [DOI] [PubMed] [Google Scholar]
- 13.He F, Li J, Xu J, Zhang S, Xu Y, Zhao W, et al. Decreased expression of ARID1A associates with poor prognosis and promotes metastases of hepatocellular carcinoma. Journal of Experimental & Clinical Cancer Research. 2015;34(1):80–88. doi: 10.1186/s13046-015-0164-3. http://doi.org/10.1186/s13046-015-0164-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, Nagl NG, Wilsker D, Van Scoy M, Pacchione S, Yaciuk P, et al. Two related ARID family proteins are alternative subunits of human SWI/SNF complexes. The Biochemical Journal. 2004;383(Pt 2):319–325. doi: 10.1042/BJ20040524. http://doi.org/10.1042/BJ20040524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Helming KC, Wang X, Wilson BG, Vazquez F, Haswell JR, Manchester HE, Kim Y, Kryukov GV, Ghandi M, Aguirre AJ, et al. ARID1B is a specific vulnerability in ARID1A-mutant cancers. Nat. Med. 2014;20:251–254. doi: 10.1038/nm.3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helming KC, Wang X, Roberts CWM. Vulnerabilities of Mutant SWI/SNF Complexes in Cancer. Cancer Cell. 2014;26(3):309–317. doi: 10.1016/j.ccr.2014.07.018. http://doi.org/10.1016/j.ccr.2014.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nature Reviews Genetics. 2014;15(4):272–286. doi: 10.1038/nrg3682. http://doi.org/10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- 18.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(50):21931–21936. doi: 10.1073/pnas.1016071107. http://doi.org/10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Phelan ML, Sif S, Narlikar GJ, Kingston RE. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Molecular Cell. 1999;3(2):247–253. doi: 10.1016/s1097-2765(00)80315-9. [DOI] [PubMed] [Google Scholar]
- 20.Saha A, Wittmeyer J, Cairns BR. Chromatin remodelling: the industrial revolution of DNA around histones. Nature Reviews Molecular Cell Biology. 2006;7(6):437–447. doi: 10.1038/nrm1945. http://doi.org/10.1038/nrm1945. [DOI] [PubMed] [Google Scholar]
- 21.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. http://doi.org/10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153(2):307–319. doi: 10.1016/j.cell.2013.03.035. http://doi.org/10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saeki N, Kuwahara Y, Sasaki H, Satoh H, Shiroishi T. Gasdermin (Gsdm) localizing to mouse Chromosome 11 is predominantly expressed in upper gastrointestinal tract but significantly suppressed in human gastric cancer cells. Mammalian Genome : Official Journal of the International Mammalian Genome Society. 2000;11(9):718–724. doi: 10.1007/s003350010138. [DOI] [PubMed] [Google Scholar]
- 24.Saeki N, Kim DH, Usui T, Aoyagi K, Tatsuta T, Aoki K, et al. GASDERMIN, suppressed frequently in gastric cancer, is a target of LMO1 in TGF-beta-dependent apoptotic signalling. Oncogene. 2007;26(45):6488–6498. doi: 10.1038/sj.onc.1210475. http://doi.org/10.1038/sj.onc.1210475. [DOI] [PubMed] [Google Scholar]
- 25.Jass JR, Biden KG, Cummings MC, Simms LA, Walsh M, Schoch E, et al. Characterisation of a subtype of colorectal cancer combining features of the suppressor and mild mutator pathways. Journal of Clinical Pathology. 1999;52(6):455–460. doi: 10.1136/jcp.52.6.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guinney J, Dienstmann R, Wang X, de Reyniès A, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nature Medicine. 2015;21(11):1350–1356. doi: 10.1038/nm.3967. http://doi.org/10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Euskirchen GM, Auerbach RK, Davidov E, Gianoulis TA, Zhong G, Rozowsky J, et al. Diverse roles and interactions of the SWI/SNF chromatin remodeling complex revealed using global approaches. PLoS Genetics. 2011;7(3):e1002008. doi: 10.1371/journal.pgen.1002008. http://doi.org/10.1371/journal.pgen.1002008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tolstorukov MY, Sansam CG, Lu P, Helming KC, Alver BH, Tillman EJ, et al. Swi/Snf chromatin remodeling/tumor suppressor complex establishes nucleosome occupancy at target promoters. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(25):10165–10170. doi: 10.1073/pnas.1302209110. http://doi.org/10.1073/pnas.1302209110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alexander JM, Hota SK, He D, Thomas S, Ho L, Pennacchio LA, Bruneau BG. Brg1 modulates enhancer activation in mesoderm lineage commitment. Development. 2015;142(8):1418–1430. doi: 10.1242/dev.109496. http://doi.org/10.1242/dev.109496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bulger M, Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011;144(3):327–339. doi: 10.1016/j.cell.2011.01.024. http://doi.org/10.1016/j.cell.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson BG, Roberts CWM. SWI/SNF nucleosome remodellers and cancer. Nature Reviews Cancer. 2011;11(7):481–492. doi: 10.1038/nrc3068. http://doi.org/10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- 32.Masliah-Planchon J, Bièche I, Guinebretière J-M, Bourdeaut F, Delattre O. SWI/SNF chromatin remodeling and human malignancies. Annual Review of Pathology. 2015;10(1):145–171. doi: 10.1146/annurev-pathol-012414-040445. http://doi.org/10.1146/annurev-pathol-012414-040445. [DOI] [PubMed] [Google Scholar]
Methods-only References
- 33.Gao X, Tate P, Hu P, Tjian R, Skarnes WC, Wang Z. ES cell pluripotency and germ-layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(18):6656–6661. doi: 10.1073/pnas.0801802105. http://doi.org/10.1073/pnas.0801802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weiser MM. Intestinal epithelial cell surface membrane glycoprotein synthesis. I. An indicator of cellular differentiation. The Journal of Biological Chemistry. 1973;248(7):2536–2541. [PubMed] [Google Scholar]
- 35.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [PMID: 20080505] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv: 1303.3997v2 [q-bio.GN] 2013 [Google Scholar]
- 37.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Research. 2012;22(3):568–576. doi: 10.1101/gr.129684.111. http://doi.org/10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnology. 2013 doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li J, Lupat R, Amarasinghe KC, Thompson ER, Doyle MA, Ryland GL, et al. CONTRA: copy number analysis for targeted resequencing. Bioinformatics (Oxford, England) 2012;28(10):1307–1313. doi: 10.1093/bioinformatics/bts146. http://doi.org/10.1093/bioinformatics/bts146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kharchenko PV, Tolstorukov MY, Park PJ. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat. Biotechnol. 2008;26:1351–1359. doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kheradpour P, Kellis M. Systematic discovery and characterization of regulatory motifs in ENCODE TF binding experiments. Nucleic Acids Res. 2014;42:2976–2987. doi: 10.1093/nar/gkt1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trapnell C, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Whole exome sequencing of tumors from MX1-Cre Arid1afl/fl mice
SWI/SNF binding sites in HCT116 cells
ChIP-qPCR data for HCT116 cells
RNA-Seq data with GSEA for HCT116 cells 14
H3K27ac regions of enrichment with nearest genes and gene ontology (GO), motif, super-enhancer (SE) analysis in HCT116 cells
ENCODE accession codes for HCT116 cells
H3K27ac regions of enrichment with nearest genes and GO, motif, SE analysis in mouse colon epithelium
RNA-Seq data with GSEA for mouse colon epithelium