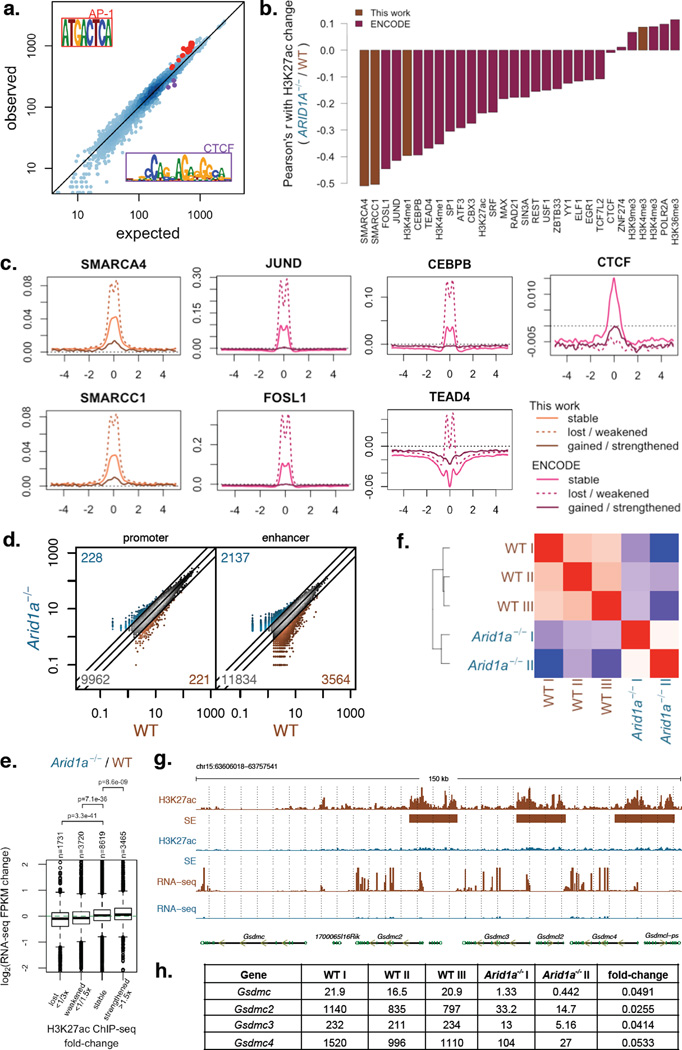
Figure 5. ARID1A loss impairs enhancer-mediated gene regulation in the colonic epithelium.
a) Observed vs. expected TF motif instances at TSS-distal H3K27ac regions (enhancers) with reduced H3K27ac signal based on enrichment regions with stable H3K27ac signal. Motifs highly similar to AP1 and CTCF motifs are highlighted;
b) Correlation between H3K27ac signal change (Arid1a−/− / WT) and WT ChIP-Seq signal levels of different factors profiled in this work and by the ENCODE Project.
c) ChIP-Seq profiles for SMARCA4, SMARCC1, JUND, FOSL1, and CTCF in WT HCT116 cells centered around TSS-distal H3K27ac regions (enhancers) that remain stable, show lost/weakened H3K27ac, or show gained/strengthened H3K27ac in Arid1a−/− cells relative to WT;
d) H3K27ac levels at TSS-proximal (promoter) and TSS-distal (enhancer) enrichment regions for colon epithelium from wildtype (WT) and Villin-CreER-T2 Arid1afl/fl (Arid1a−/−) mice (Note: numbers in the three corners denote numbers of activated (>2×), inactivated (<1/2×) and stable sites);
e) Fold changes (log2) of gene expression between WT and Arid1a−/− mouse colon epithelium for genes nearest to TSS-distal H3K27ac regions (enhancers) split based on Arid1a−/− / WT ChIP-Seq signal;
f) Pearson's correlation among RNA-Seq samples based on FPKM values for mouse colon epithelium dissociated from individual wildtype mice (WT, n=3) and Villin-CreER-T2 Arid1afl/fl mice (Arid1a−/−, n=2);
G) H3K7ac ChIP-Seq tracks, super-enhancer (SE) calls, and RNA-Seq tracks at Gsdmc locus in WT and Arid1a−/− mouse colon epithelium.
H) RNA-Seq FKPM values for individual WT and Arid1a−/− mice for Gsdmc, Gsdmc2, Gsdmc3, and Gsdmc4.