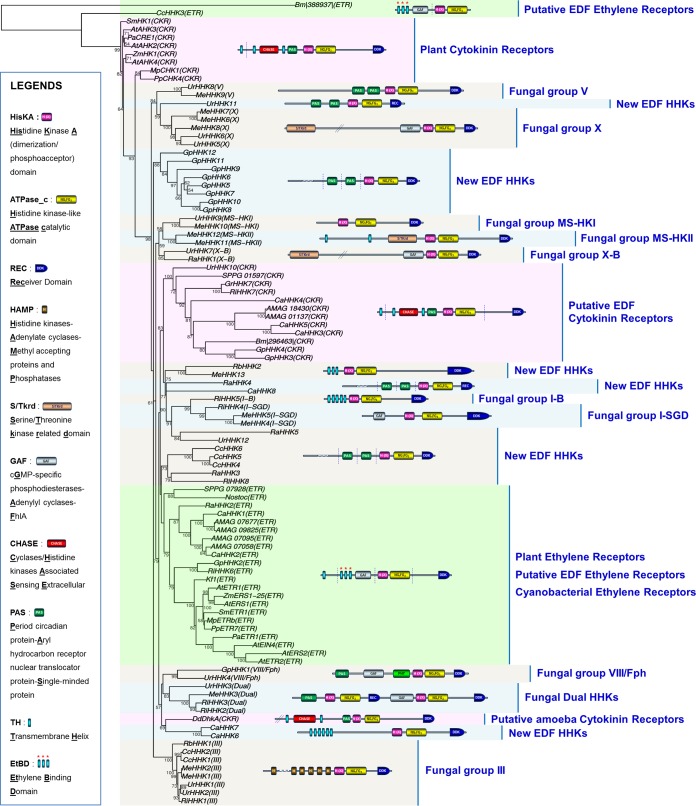
FIG 5 .
Phylogeny estimation of HHK predicted protein sequences. Methods used to carry out this phylogenetic analysis are provided in Text S1 in the supplemental material. Early diverging fungi predicted HHK sequences were categorized following sequence analysis of both HisKA signatures and N-terminal sensing domains according to previous classifications (4, 10). Abbreviations: Ri, Rhizophagus irregularis (Glomeromycotina); Me, Mortierella elongata (Mortierellomycotina); Ur, Umbelopsis ramanniana (Mucoromycotina); Cc, Conidiobolus coronatus (Entomophthoromycotina); Rb, Ramicandelaber brevisporus (Kickxellomycotina); Ca, Catenaria anguillulae (Blastocladiomycota); Gp, Gonapodya prolifera (Chytridiomycota); Ra, Rozella allomycis (Cryptomycota); AMAG, Allomyces macrogynus (Blastocladiomycota); Dd, Dictyostelium discoideum (Amoebae); SPPG, Spizellomyces punctatus (Chytridiomycota); Bm, Basidiobolus meristoporus (Entomophthoromycotina); Gr, Gigaspora rosea (Glomeromycotina); At, Arabidopsis thaliana (dicots); Zm, Zea mays (monocots); Pa, Picea abies (gymnosperms); Sm, Selaginella moellendorffii (lycophytes); Pp, Physcomitrella patens (bryophytes); Mp, Marchantia polymorpha (liverworts); Kf, Klebsormidium flaccidum (charophytes).