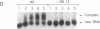Abstract
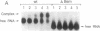
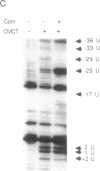
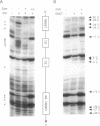
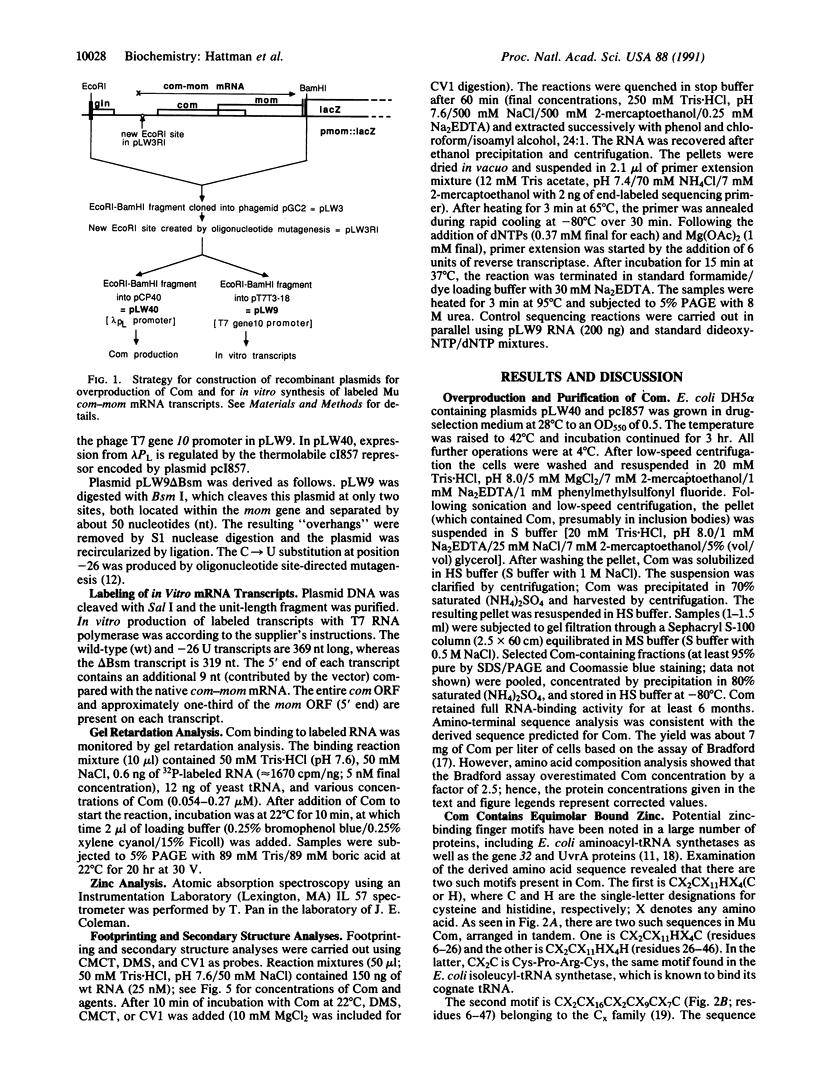
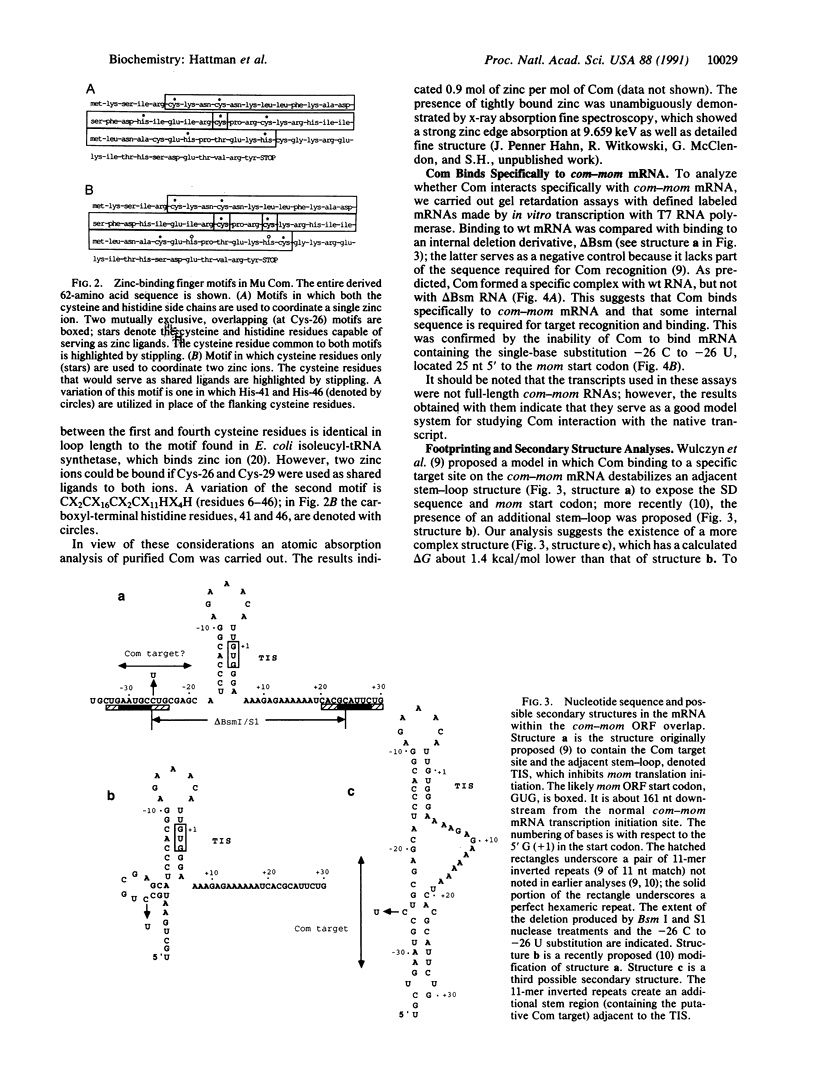
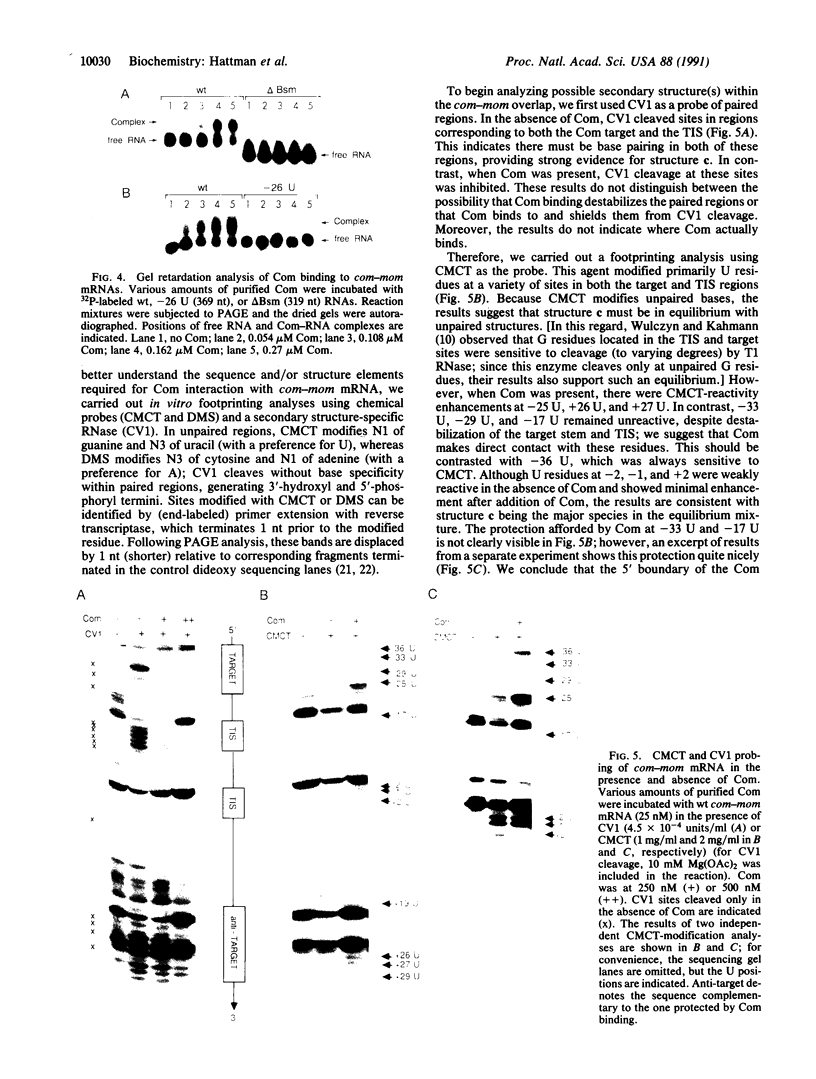
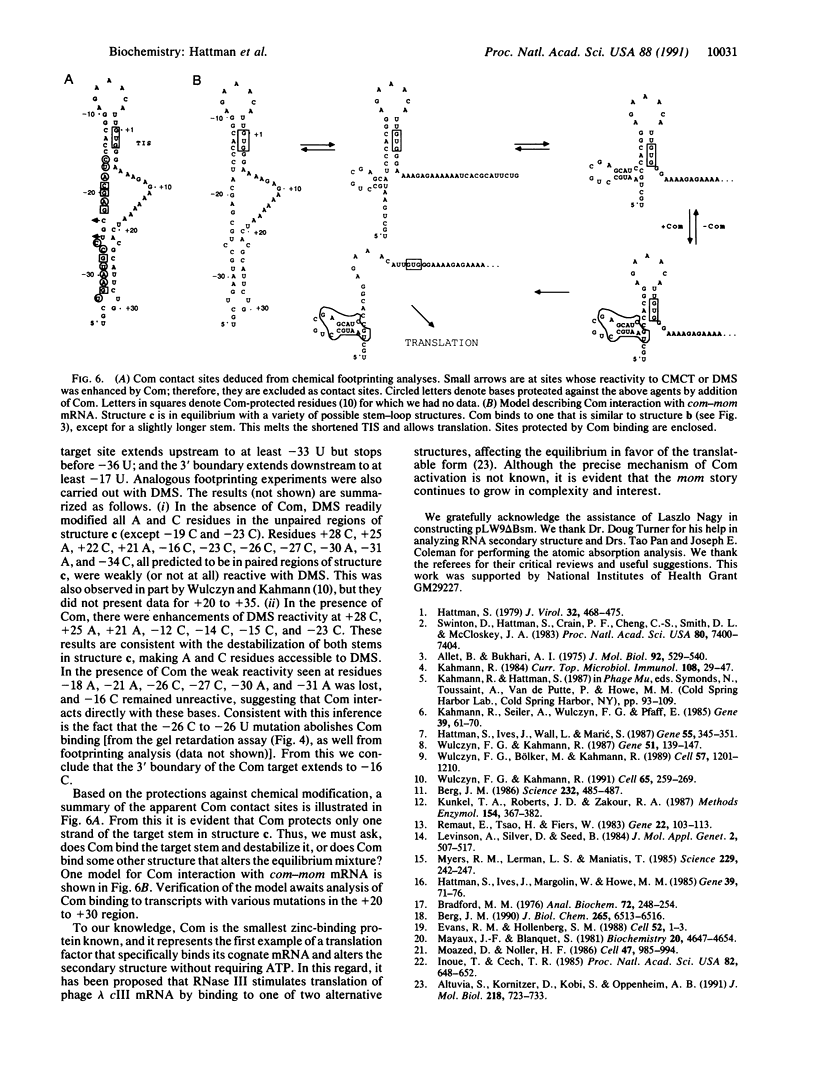
Bacteriophage Mu controls an unusual DNA-modification function encoded by the mom gene, which is located in an operon that consists of two overlapping genes. The com gene, located proximal to the 5' end of the common mRNA transcript, encodes a polypeptide of 62 amino acids that is required for translation of mom. Analysis of the derived amino acid sequence reveals that Com contains zinc-binding finger motifs, suggesting that Com may be a zinc-activated regulatory protein. Atomic absorption analysis showed that there is about one zinc bound per molecule of Com. We have subcloned the com gene into an expression vector and thus have overproduced and purified the Com protein. By gel retardation analysis with various 32P-labeled RNAs (made by in vitro transcription with T7 RNA polymerase), we show that Com binds specifically to com-mom mRNA. A single C----U substitution mutation, located 26 nucleotides upstream from the mom translation start codon, abolishes Com binding. The nature of the Com target sequence was deduced from in vitro footprinting analyses. The results are consistent with the existence of a complex stem-loop structure within the overlap of the com-mom open-reading-frames. Com binding to its target site results in the destabilization of a proposed translation-inhibitor stem-loop (TIS) to expose the Shine-Dalgarno sequence and mom translation initiation codon. This suggests that Com interaction with a specific site on its cognate mRNA alters the mRNA secondary structure to activate translation of mom.
Full text
PDF
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allet B., Bukhari A. I. Analysis of bacteriophage mu and lambda-mu hybrid DNAs by specific endonucleases. J Mol Biol. 1975 Mar 15;92(4):529–540. doi: 10.1016/0022-2836(75)90307-1. [DOI] [PubMed] [Google Scholar]
- Altuvia S., Kornitzer D., Kobi S., Oppenheim A. B. Functional and structural elements of the mRNA of the cIII gene of bacteriophage lambda. J Mol Biol. 1991 Apr 20;218(4):723–733. doi: 10.1016/0022-2836(91)90261-4. [DOI] [PubMed] [Google Scholar]
- Berg J. M. Potential metal-binding domains in nucleic acid binding proteins. Science. 1986 Apr 25;232(4749):485–487. doi: 10.1126/science.2421409. [DOI] [PubMed] [Google Scholar]
- Berg J. M. Zinc fingers and other metal-binding domains. Elements for interactions between macromolecules. J Biol Chem. 1990 Apr 25;265(12):6513–6516. [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Evans R. M., Hollenberg S. M. Zinc fingers: gilt by association. Cell. 1988 Jan 15;52(1):1–3. doi: 10.1016/0092-8674(88)90522-3. [DOI] [PubMed] [Google Scholar]
- Hattman S., Ives J., Margolin W., Howe M. M. Regulation and expression of the bacteriophage mu mom gene: mapping of the transactivation (dad) function to the C region. Gene. 1985;39(1):71–76. doi: 10.1016/0378-1119(85)90109-x. [DOI] [PubMed] [Google Scholar]
- Hattman S., Ives J., Wall L., Marić S. The bacteriophage Mu com gene appears to specify a translation factor required for mom gene expression. Gene. 1987;55(2-3):345–351. doi: 10.1016/0378-1119(87)90295-2. [DOI] [PubMed] [Google Scholar]
- Hattman S. Unusual modification of bacteriophage Mu DNA. J Virol. 1979 Nov;32(2):468–475. doi: 10.1128/jvi.32.2.468-475.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T., Cech T. R. Secondary structure of the circular form of the Tetrahymena rRNA intervening sequence: a technique for RNA structure analysis using chemical probes and reverse transcriptase. Proc Natl Acad Sci U S A. 1985 Feb;82(3):648–652. doi: 10.1073/pnas.82.3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahmann R., Seiler A., Wulczyn F. G., Pfaff E. The mom gene of bacteriophage mu: a unique regulatory scheme to control a lethal function. Gene. 1985;39(1):61–70. doi: 10.1016/0378-1119(85)90108-8. [DOI] [PubMed] [Google Scholar]
- Kahmann R. The mom gene of bacteriophage Mu. Curr Top Microbiol Immunol. 1984;108:29–47. doi: 10.1007/978-3-642-69370-0_4. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Levinson A., Silver D., Seed B. Minimal size plasmids containing an M13 origin for production of single-strand transducing particles. J Mol Appl Genet. 1984;2(6):507–517. [PubMed] [Google Scholar]
- Mayaux J. F., Blanquet S. Binding of zinc to Escherichia coli phenylalanyl transfer ribonucleic acid synthetase. Comparison with other aminoacyl transfer ribonucleic acid synthetases. Biochemistry. 1981 Aug 4;20(16):4647–4654. doi: 10.1021/bi00519a020. [DOI] [PubMed] [Google Scholar]
- Moazed D., Noller H. F. Transfer RNA shields specific nucleotides in 16S ribosomal RNA from attack by chemical probes. Cell. 1986 Dec 26;47(6):985–994. doi: 10.1016/0092-8674(86)90813-5. [DOI] [PubMed] [Google Scholar]
- Myers R. M., Lerman L. S., Maniatis T. A general method for saturation mutagenesis of cloned DNA fragments. Science. 1985 Jul 19;229(4710):242–247. doi: 10.1126/science.2990046. [DOI] [PubMed] [Google Scholar]
- Remaut E., Tsao H., Fiers W. Improved plasmid vectors with a thermoinducible expression and temperature-regulated runaway replication. Gene. 1983 Apr;22(1):103–113. doi: 10.1016/0378-1119(83)90069-0. [DOI] [PubMed] [Google Scholar]
- Swinton D., Hattman S., Crain P. F., Cheng C. S., Smith D. L., McCloskey J. A. Purification and characterization of the unusual deoxynucleoside, alpha-N-(9-beta-D-2'-deoxyribofuranosylpurin-6-yl)glycinamide, specified by the phage Mu modification function. Proc Natl Acad Sci U S A. 1983 Dec;80(24):7400–7404. doi: 10.1073/pnas.80.24.7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulczyn F. G., Bölker M., Kahmann R. Translation of the bacteriophage Mu mom gene is positively regulated by the phage com gene product. Cell. 1989 Jun 30;57(7):1201–1210. doi: 10.1016/0092-8674(89)90057-3. [DOI] [PubMed] [Google Scholar]
- Wulczyn F. G., Kahmann R. Post-transcriptional regulation of the bacteriophage Mu mom gene by the com gene product. Gene. 1987;51(2-3):139–147. doi: 10.1016/0378-1119(87)90302-7. [DOI] [PubMed] [Google Scholar]
- Wulczyn F. G., Kahmann R. Translational stimulation: RNA sequence and structure requirements for binding of Com protein. Cell. 1991 Apr 19;65(2):259–269. doi: 10.1016/0092-8674(91)90160-z. [DOI] [PubMed] [Google Scholar]