Figure 6. GapR localization during the cell cycle is correlated with replisome dynamics.
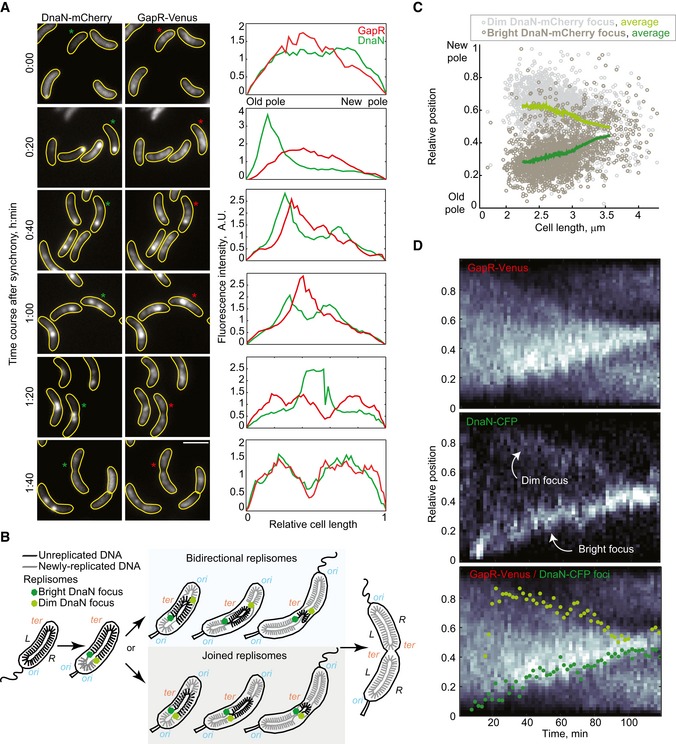
- Images showing GapR‐Venus and DnaN‐mCherry localization in a time‐course experiment (CJW5744) following cell cycle synchronization. After synchronization, cells were resuspended in M2G liquid medium and incubated at 30°C, and samples were taken for imaging every 20 min and imaged. Fluorescence intensity profiles along the cell length are shown for cells marked with an asterisk. Scale bar = 2 μm.
- Schematic showing the organization of the unreplicated and replicated chromosomal DNA coordinated with replisome dynamics during the Caulobacter crescentus cell cycle. The two replisomes move toward midcell, either in a joined fashion from the old pole or bidirectionally from opposite poles. The origin (ori) and terminus (ter) of replication as well as the left (L) and right (R) arms of the chromosome are indicated.
- Relative locations of dim and bright DnaN‐mCherry foci in cells exhibiting two foci. See Code EV1 information for details of focus detection algorithm.
- Kymographs of single‐cell time‐lapse microscopy experiment. After synchronization, swarmer cells (CJW5836) were spotted into a 1% agarose M2G pad at ~25° and imaged every 2 min. The bottom panel shows the kymograph of GapR‐Venus overlaid with the position of the dim (light green) and bright (dark green) DnaN‐CFP foci.
