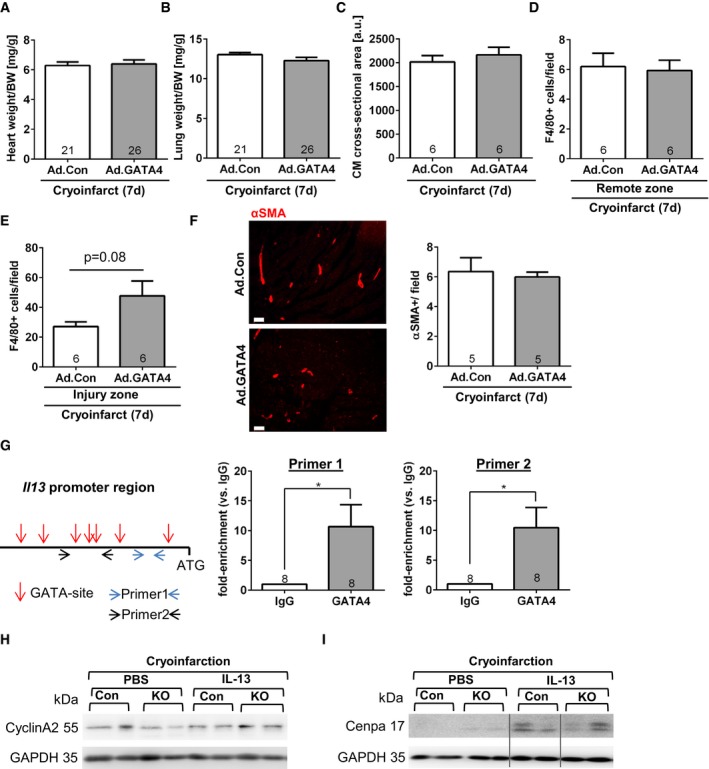
Heart weight/body weight (BW) ratio of the indicated mice 7 days after sham surgery or cryoinfarction and application of adenoviruses as shown.
Lung weight/body weight ratio of the indicated mice 7 days after sham surgery or cryoinfarction and application of adenoviruses as shown.
Cardiomyocyte (CM) cross‐sectional area assessed 7 days after cryoinfarction in the indicated mice.
Quantification of myocardial macrophage abundance (positive for F4/80) remote of the infarcted area of the indicated mice.
Quantification of myocardial macrophage abundance (positive for F4/80) within the infarcted area of the indicated mice.
Myocardial immunofluorescence staining for αSMA in mice 7 days after cryoinjury and treatment with the indicated viruses. Quantification of small αSMA‐positive small conductance vessel (˜20–50 μm in diameter) in the myocardium of mice as shown. Scale bars: 50μm.
Scheme of the mouse Il13 promoter region (˜700 bp) before the ATG translational start sites, GATA binding regions are marked by a red arrow, and the primers used are indicated by blue or black arrows. On the left, the results of a chromatin immunoprecipitation assay from neonatal hearts 7 days after cryoinjury and after immunoprecipitation with control IgG or GATA4 are shown. *P = 0.0197 for primer 1 and *P = 0.0142 for primer 2.
Representative immunoblots for cyclin A2 and GAPDH from hearts of the mice as indicated (con = control, KO = CM‐G4‐KO).
Representative immunoblots for cenpa and GAPDH from hearts of the mice as indicated. All lanes were run on the same gel but were non‐contiguous where indicated by the gray lines.
Data information: (A–G) The number within bars indicates the number of mice analyzed in that particular group. All data are expressed as mean ± SEM. Unpaired Student's
‐test was used to compare groups.