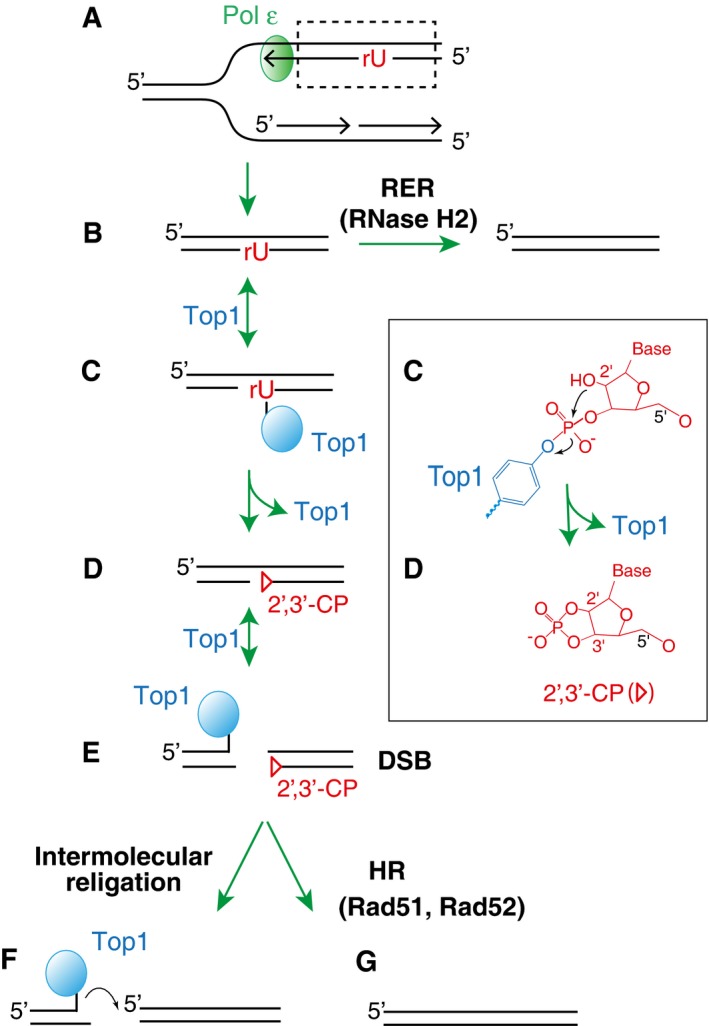
Ribonucleotides are preferentially incorporated on the leading strand when Pol ε is genetically altered.
Newly incorporated ribonucleotides are normally removed by RER initiated by RNase H2 incision at the ribonucleotide site.
In the absence of RNase H2, Top1 forms a cleavage complex at the ribonucleotide site.
The attack by 2′‐hydroxyl group on the phosphotyrosyl bond releases Top1, converting the ribonucleotides into nicks with 2′,3′‐cyclic phosphate ends (triangle). A detailed biochemical reaction is shown in the box on the right.
Subsequent cleavage by Top1 on the opposite strand of the existing nick leads to a DSB.
The resulting DSB with covalently linked Top1 at the end is prone to religate/recombine with other DNA ends.
Alternatively, homologous recombination (dependent on Rad51 and Rad52) repairs the DSB.