Figure EV5. Scheme of the different outcomes resulting from Top1 activity at incorporated ribonucleotides.
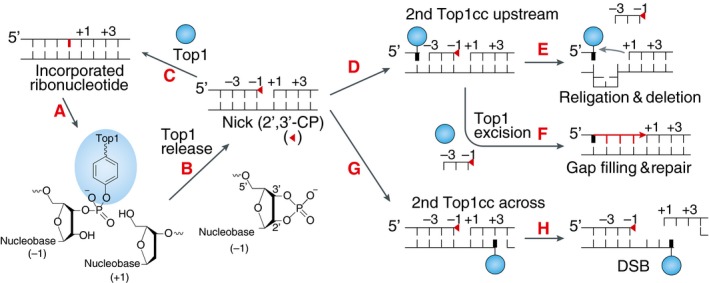
- Following ribonucleotide incorporation into the DNA (shown in red) during replication, a Top1cc forms at a ribonucleotide site.
- Top1cc is reversed by the nucleophilic attack of the 2′‐ribose hydroxyl, which generates a 2′,3′‐cyclic phosphate (2′,3′‐CP) end (red triangle) and releases catalytically active Top1.
- Top1 can then bind to the nick, enabling nucleophilic attack of the 5′‐ribose hydroxyl on 2′,3′‐CP and facilitating religation of DNA backbone independently of its enzymatic activity (Huang et al, 2015).
- Sequential Top1 cleavage on the same strand, upstream of the 2′,3′‐CP by the released Top1 or by another Top1 can lead to the loss of short DNA fragment containing 2′,3′‐CP and persistent Top1cc.
- Alternatively, endonucleolytic cleavage (not shown) or excision of the Top1cc by tyrosyl‐DNA phosphodiesterase 1 (TDP1) (Sparks & Burgers, 2015) followed by gap filling can repair the DNA while eliminating the ribonucleotide.
- When the sequential Top1 cleavage is on the opposite strand to the 2′,3′‐CP, a DSB is formed.
- Such Top1‐linked DSBs can cause replication stress and lead to recombinations. Adapted from Pommier et al (2016).
