Figure EV3. An exchange of Met and Ser residues in Oct4 and Oct6 POU domains is responsible for different DNA‐binding preferences.
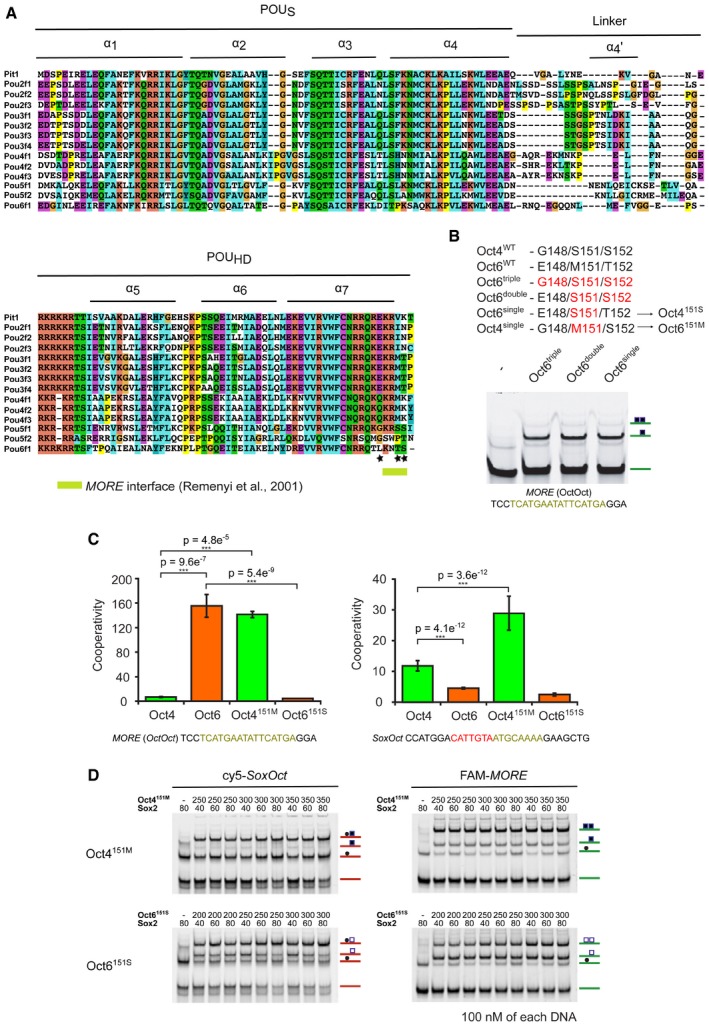
- Sequence alignment of POU domains of Oct family TFs. POUS, the linker region, and POUHD, as well as individual helixes, are labeled. Notably, this structure‐based alignment differs slightly from the one used previously 44, which is explained by a low conservation of residues in the N‐terminal part of the POU linker. The alignment was prepared in T‐Coffee software and colors distinguish conservation and amino acid residue types. A protein surface interacting with the MORE DNA (as shown in 17) is highlighted. Each of three studied residues (B) is marked by a black asterisk.
- An overview of tested Oct4 and Oct6 mutants with amino acid exchanges in their POUHD. The EMSA below shows the low cooperativity on MORE DNA element caused by the mutations. As the single mutation of the 151 site had the same effect as double and triple mutations, the single mutation was selected for further study.
- Bar plots of EMSA‐derived cooperativity factors show different preferences between formation of Oct–Oct homodimers on MORE (left) and Sox2–Oct heterodimers on the SoxOct element (right) for WT and mutated forms of Oct4 and Oct6 POU domains. Cooperativity values for WT Oct4 and Oct6 correspond to the values shown in Fig 1E and F and are shown again for comparison with the mutant proteins. The mean is shown with standard deviation as error bars (n = 3–13), and Tukey's multiple comparison of means was performed to assess statistical significance (***P < 0.001).
- Representative EMSAs performed in single‐tube reactions containing the Sox2 HMG and Oct4151M and Oct6151S POU domains of Oct transcription factors. Related to the quantification in Fig 2D.
