Abstract
Many plasmids used for gene cloning and heterologous protein expression in E. coli cells are low copy number or single copy number plasmids. Extraction of these types of plasmids from small bacterial cell cultures produces low DNA yields. In this study we have quantitated yields of low copy and single copy number plasmid DNAs after growth of cells in four widely used broths (SB, SOC, TB and 2xYT) and compared results to those obtained with LB, the most common E. coli cell growth medium. TB (terrific broth) consistently generated the greatest amount of plasmid DNA, in agreement with its ability to produce higher cell titers. The superiority of TB was primarily due to its high levels of yeast extract (24 g/L) and was independent of glycerol, a unique component of this broth. Interestingly, simply preparing LB with similarly high levels of yeast extract (LB24 broth) resulted in plasmid yields that were equivalent to those of TB. In contrast, increasing ampicillin concentration to enhance plasmid retention did not improve plasmid DNA recovery. These experiments demonstrate that yields of low and single copy number plasmid DNAs from minipreps can be strongly enhanced using simple and inexpensive media.
Keywords: plasmid, copy number, DNA purification, miniprep, TB, LB
1. Introduction
Molecular biologists routinely use circular plasmids propagated in Escherichia coli cells for cloning, expression, sequencing and mutagenesis of genes. These plasmids are commonly classified into three groups – high copy number (corresponding to hundreds of plasmid DNA molecules per cell), low copy number (typically less than fifty plasmids per cell), and single copy number (approximately one per cell) (Summers, 1998; Feinbaum, 2002; Nordstrom and Dasgupta, 2006). Most vectors used for simple DNA fragment cloning are high copy number plasmids. Extraction of such plasmids from E. coli cells produces high yields and the DNAs are frequently purified from 1.5 mL of overnight cultures of bacterial cells (referred to as plasmid minipreps). In contrast, many vectors used for expression of proteins are low copy number plasmids. Having reduced copy number (lower gene dosage) can decrease basal, or uninduced, intracellular levels of an expressed protein, which is advantageous because some foreign proteins are toxic even at very low levels and therefore can inhibit growth of the E. coli cells. Examples of such plasmids include the widely used pET series of vectors (Novagen), IMPACT system vectors (New England Biolabs), HaloTag vectors (Promega), and several others (Dersch et al., 1994; Mardanov et al., 2007; Cheong et al., 2013; Guan et al., 2013). Yields of these plasmids are greatly reduced compared to high copy number plasmids and therefore larger cell cultures must be inoculated in order to extract sufficient amounts of DNA for detection and manipulation. Plasmids employed for some other procedures such as genomic DNA cloning and sequencing are also low copy or even single copy number plasmids and therefore also produce low DNA yields upon extraction.
The most commonly used liquid medium for growth of E. coli cells is LB (Lysogeny Broth), but several other broths have been developed over the years and employed for specific procedures. Examples of these other media include SB (Super Broth), SOC (Super Optimal broth with Catabolite repression), TB (Terrific Broth), and 2xYT (Yeast extract Tryptone) (Tartoff and Hobbs, 1987; Elbing and Brent, 2002; Green and Sambrook, 2012; Lessard, 2013). SOC is added to E. coli cells to aid recovery after exposure to high salt concentrations and heat shock in many chemical-based DNA transformation protocols and 2xYT is used in many bacteriophage infection protocols (Elbing and Brent, 2002; Green and Sambrook, 2012). All of these growth media share two components, tryptone and yeast extract, but the amounts of the two nutrients within each broth are variable. Some of the broths contain specific additional chemicals that are beneficial to cell growth such as glycerol, glucose and/or salts.
In the current study, we have tested five well established E. coli broths to determine which one produces the highest yields of low copy and single copy number plasmid DNAs from 1.5 mL minipreps. TB broth consistently produced the greatest DNA yields. The strong results with TB were found to be independent of its glycerol content and, instead, were primarily due to its high concentration of yeast extract. Adding increasing amounts of yeast extract to LB broth produced higher DNA yields in a concentration-dependent manner. The results demonstrate that yields of both low and single copy number plasmids isolated using standard alkaline lysis minipreps can be strongly enhanced using simple, inexpensive media containing elevated levels of yeast extract.
2. Materials and methods
2.1. Strains, plasmids and reagents
E. coli Top10 cells containing either the low copy number plasmid pET15b-rKGA (Farrell and Taylor, 2005), the single copy number plasmid pBeloBAC11 (Tillett and Neilan, 1998) (obtained from New England Biolabs), or the high copy number plasmid pRS425 (Christianson et al., 1992) were used for all experiments. Yeast extract (VWR 90004-092) was produced by BD Biosciences and tryptone (VWR 97063-424) was from Amresco. Ampicillin (VWR IB02040) was sold by IBI Scientific. All broths were prepared according to the descriptions published in Current Protocols in Molecular Biology (Elbing and Brent, 2002). Recipes used to prepare 1 L of each broth were the following:
LB: 10 g tryptone, 5 g yeast extract, 5 g NaCl, 1 mL 1 M NaOH.
LB24: 10 g tryptone, 24 g yeast extract, 5 g NaCl, 1 mL 1 M NaOH.
TB: 12 g tryptone, 24 g yeast extract, 4 mL glycerol, 100 mL 0.17 M KH2PO4/0.72 M K2HPO4.
SB: 32 g tryptone, 20 g yeast extract, 5 g NaCl, 5 mL 1 M NaOH.
2xYT: 16 g tryptone, 10 g yeast extract, 5 g NaCl.
SOC: 20 g tryptone, 5 g yeast extract, 0.58 g NaCl, 3.6 g glucose, 2.5 mM KCl, 10 mM MgCl2, 10 mM MgSO4.
2.2. DNA fluorometry, gel electrophoresis and cell culture density determination
Cell culture densities were monitored by measuring light scattering at 550 nm using a BioRad Smartspec spectrophotometer after diluting overnight cultures 1:20 into water. DNA fluorometry was performed using a Qubit 2.0 fluorometer (Life Technologies-Invitrogen) in conjunction with the broad range assay protocol. Electrophoretic analysis of purified DNAs employed Horizon 11–14 rigs (Labrepco), 1 x TAE electrophoresis buffer and 0.7–0.8% agarose gels run at approximately 130 volts. Gels were stained with ethidium bromide and photographs were taken using an Alpha Innotech RED instrument.
2.3. Plasmid DNA miniprep protocols
Most plasmid minipreps were performed using a standard alkaline lysis protocol (Elbing and Brent, 2002; Green and Sambrook, 2012) in conjunction with 2–4 mL cell cultures containing 100 μg/ml ampicillin that were shaken overnight (20–22 hours) at ~ 250 rpm at 37°C in glass tubes (20x150 - VWR 47729-584) or plastic snapcap tubes (17x100 - VWR 60818-725). Briefly, standard alkaline lysis minipreps involved pelleting of 1.5 mL cells in a microcentrifuge for 20 seconds followed by addition of cold Solution I (50 mM glucose, 25 mM Tris [pH 8.0], 10 mM EDTA), fresh Solution II (0.2 M NaOH, 1% SDS), and cold Solution III (3 M KOAc). After proteins and cell debris were sedimented for 10 min at 21,000g, supernatants were transferred to a new tube, removing only the liquid that was ~ 4 mm above the protein pellet to minimize chromosomal DNA contamination. The DNA was precipitated by addition of 0.4 mL isopropanol and centrifugation for 5 min. After washing the DNA pellets with 0.5 mL cold 70% ethanol, they were dried in a Savant DNA120 speedvac for 10 minutes and resuspended in 50 μL TE (10 mM Tris [pH 8.0], 1 mM EDTA) plus 2 μL 2 mg/mL RNase A. In some experiments Qiagen miniprep kits (#27104) containing anion exchange spin columns were employed.
For experiments involving multiple broths, all overnight cultures were inoculated with the same initial number of E. coli cells and shaken together at 37°C overnight. Typically, this involved harvesting cells from the surface of an LB + ampicillin plate into a microfuge tube containing 0.5 mL LB broth, vortexing, and subsequently aliquotting 20 μl cells into each cell culture tube. Four or five cell cultures were initiated for each broth. After DNA concentrations and cell titers were calculated, the resulting averages and standard deviations are shown in the figures. For tests involving the single copy number plasmid pBeloBAC11, cells were grown in media containing 20 μg/ml chloramphenicol.
3. Results and discussion
3.1. Evaluation of plasmid DNA yields using five common broths
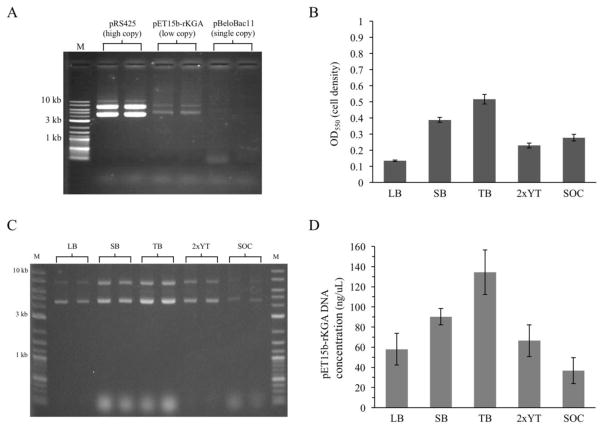
The primary goal of this project was to identify broths and/or growth conditions that enhance yields of low copy and single copy number plasmids from 1.5 mL minipreps. The specific problem that was addressed is illustrated in Figure 1A. DNAs were purified from 1.5 mL LB broth + ampicillin overnight cultures of Top10 cells containing pRS425, pET15b-rKGA, or pBeloBAC11 (high, low and single copy number plasmids, respectively) and 2 μl out of 50 total μl was mixed with loading dye and run on a 0.7% agarose gel. The two high copy number plasmid preps produced intense bands at 4 kb and 6 kb, corresponding to the supercoiled and nicked open circular forms of the plasmid. In contrast, faint bands are present in the lanes corresponding to the low and single copy number plasmids, demonstrating that cells grown in LB broth generate poor yields with these types of DNAs.
Fig. 1.
Assessment of cell culture densities and yields from 1.5 mL plasmid minipreps after propagation in five common broths. (A) Use of the most common broth, LB, produces generous amounts of high copy number DNAs, but not low or single copy number plasmids. Two μL of each 50 μL prep were loaded onto a 0.7% agarose gel. Results from two independent cultures are shown for each plasmid. (B) TB broth cultures had the highest cell densities after shaking overnight at 37°C. (C and D) Electrophoresis and DNA fluorometry of plasmid DNAs extracted from each overnight culture by alkaline lysis. Results in B–D are averages of 4 independent cultures. Error bars indicate standard deviations.
The potential for other well known broths to improve yield of the low copy number plasmid pET15b-rKGA was investigated next. Cell culture densities and plasmid DNA yields were measured for cultures shaken overnight at 37°C in LB, SB, TB, 2xYT or SOC broths containing ampicillin. All broths were inoculated with the same number of cells. TB broth consistently produced both the highest cell densities (based on OD550 readings resulting from light scattering) and the highest DNA concentrations (Figure 1B and 1D). Cell densities achieved with TB were over 3 times greater than those seen with the most commonly used broth, LB, and DNA yields were 2.5 fold higher with nonoverlapping standard deviations (Figure 1B and 1D). This result was also apparent when an aliquot of each prep was run on an agarose gel, where SB, TB and 2xYT all outperformed LB broth (Figure 1C). SOC, the only broth containing supplemental glucose, consistently produced the lowest DNA yields (Figure 1C and 1D), despite the fact that growth of cells in this broth was enhanced compared to LB (Figure 1B).
3.2. High plasmid DNA yields obtained with TB are not due to glycerol content
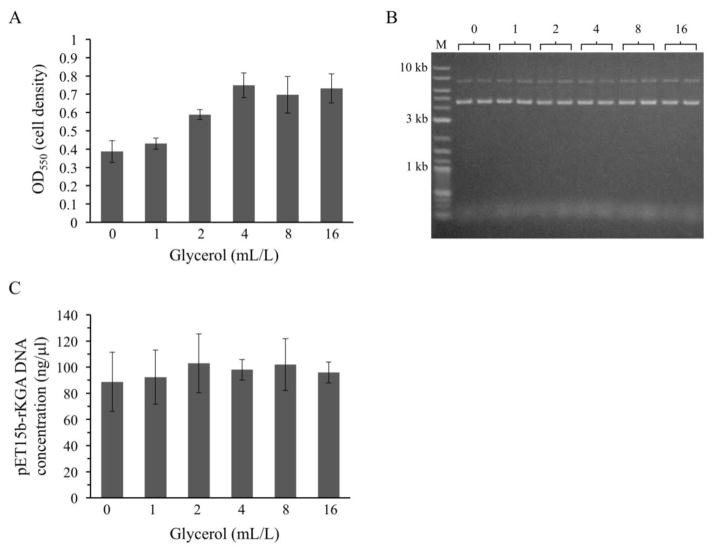
The best broth, TB, differs from the others in that it contains 4 mL glycerol per liter (see recipes in Methods), which is considered an important additional carbon source for cell growth in this medium (Tartoff and Hobbs, 1987; da Silva et al., 2009; Lessard, 2013). The importance of glycerol was tested by preparing TB media with varying amounts of glycerol, inoculating each broth with equivalent numbers of pET15b-rKGA-containing cells, and assessing cell growth and plasmid DNA yields. Although cell densities were modestly increased at 4, 8 and 16 mL per liter glycerol concentrations (Figure 2A), the quantity and quality of the resulting purified plasmid DNAs were not dependent on the presence of glycerol (note the similar band intensities in Figure 2B and the overlapping standard deviations in the fluorometry measurements in Figure 2C).
Fig. 2.
Yields of the low copy number plasmid (pET15b-rKGA) in TB broth supplemented with different concentrations of glycerol. (A) Cell titers are increased modestly at higher concentrations of supplemental glycerol. TB normally contains 4 mL/L glycerol. (B and C) The quantity and quality of plasmid DNA extracted from TB cultures is independent of glycerol concentration. Results from two separate cultures are shown in (B).
3.3. The level of yeast extract affects cell culture densities and plasmid DNA yields
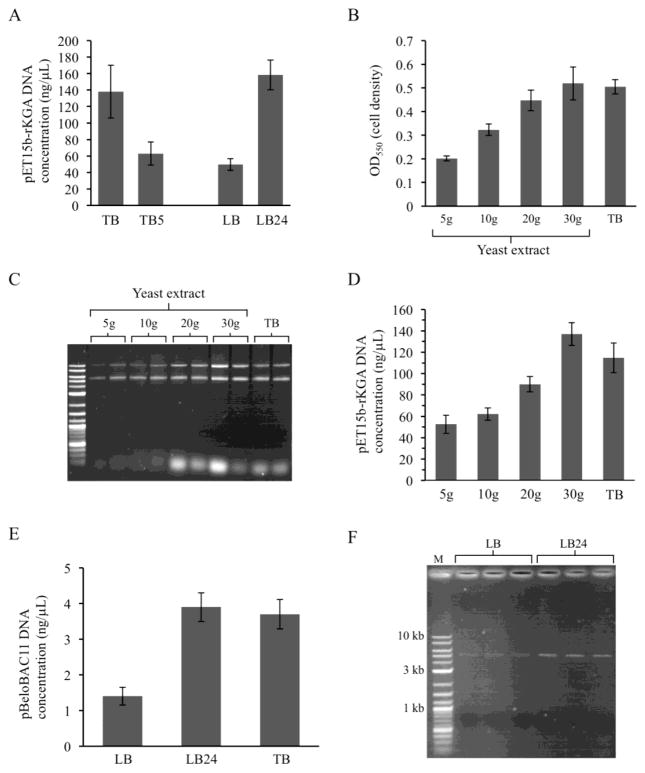
Another difference between TB and LB is that TB has 24 g/L of yeast extract versus only 5 g/L in LB broth. The impact of this nutrient was assessed initially by comparing DNA yields in (a) TB versus TB prepared normally but with only 5 g/L yeast extract (TB5) and (b) LB versus LB prepared with 24 g/L yeast extract (LB24). As shown in Figure 3A, reducing yeast extract in TB to that found in LB broth decreased DNA yields, while increasing the amount of yeast extract in LB to that seen in TB strongly increased DNA recovery. Dependence on this nutrient was investigated further by preparing LB broth with yeast extract concentrations of 5, 10, 20 and 30 g/L and performing alkaline lysis minipreps with pET15b-rKGA as before. The amount of DNA that could be extracted was found to be critically dependent upon yeast extract concentration. Both cell culture densities (Figure 3B) and DNA yields (Figure 3C and 3D) were low with 5 g/L and 10 g/L, but addition of 20 g/L or 30 g/L yeast extract produced levels that were equivalent to those seen with TB broth, with yields increased by 2.7 fold at 30 g/L.
Fig. 3.
Impact of increasing yeast extract concentrations on yields of low and single copy number plasmids. (A) Direct comparison of pET15b-rKGA DNA yields using TB, TB with only as much yeast extract as LB (5 g; called TB5), LB, and LB containing as much yeast extract as TB (called LB24). (B–D) Cell culture densities and DNA yields from low copy number plasmid pET15b-rKGA minipreps performed using LB broth containing varying amounts of yeast extract (5 g/L is normal). (E and F) Purification of single copy number plasmid pBeloBAC11 using high yeast extract broths TB and LB24 increases DNA yields. Because of the smaller amounts of DNA used, the gel in (F) was stained with ethidium bromide for twice as long as the other gels (30 min).
3.4. High yeast extract broths permit isolation of practical amounts of single copy plasmid DNA from minipreps
Purification of single copy number plasmids results in even lower yields than low copy plasmids (Figure 1A), and it is common to perform midipreps (i.e., using ~ 100 mL of cells or more) rather than minipreps to isolate these types of plasmids. We tested the ability of broth with a high level of yeast extract to enhance recovery of these plasmids from small cultures. Top10 cells containing pBeloBAC11 (Camr) were grown overnight in broths containing chloramphenicol as before. We have observed previously that minipreps performed using Qiagen miniprep kits typically produce plasmid DNA preparations with a low amount of the nicked open circular form (upper band) visible on gels such that more molecules are concentrated into a single supercoiled DNA band. This is advantageous for visualization of small amounts of DNA after staining with ethidium bromide because all of the staining is concentrated into one intense band rather than being split into two weaker bands. pBeloBAC11 DNAs purified from 1.5 mL overnight cultures using Qiagen kits were analyzed by fluorometry and gel electrophoresis. Yields of these DNAs were considerably lower than with the low copy number plasmids but, importantly, TB and LB24 generated three times as much DNA as LB broth (Figure 3E). Electrophoresis of DNAs isolated from three low and three high yeast extract cultures confirmed the purity and yield enhancement in high yeast extract broth (Figure 3F). Note that 10 μL of each 50 μL prep were loaded onto the gel shown in (F), which is more than was used for the previous gels. Use of DNA stains with greater sensitivity than ethidium bromide such as SYBR Gold (Molecular Probes) or GelRed (Biotium) would permit smaller volumes to be used for electrophoresis, thereby improving the analysis of single copy plasmids even further.
3.5. Increasing selective pressure for plasmid retention within cells does not improve yields of low copy number plasmid DNA
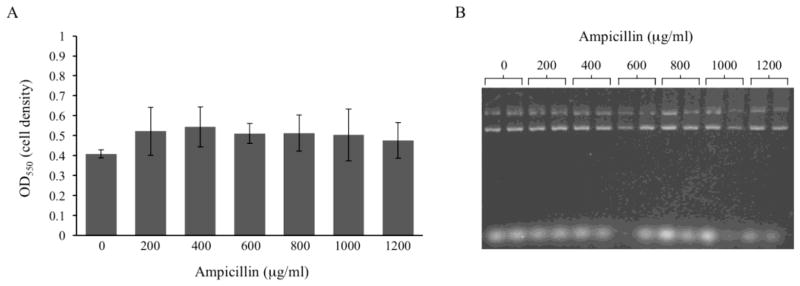
Some molecular biology laboratory technique websites have suggested that increasing the amount of antibiotic present in E. coli cell cultures, which increases selective pressure for retention of the plasmids, can enhance DNA miniprep yields. We were not able to identify a peer-reviewed source supporting this contention and therefore tested the hypothesis using the low copy number plasmid pET15b-rKGA (Ampr). Top10 cells containing the plasmid were harvested from colonies on standard LB plates containing 100 μg/mL ampicillin and were inoculated into overnight cultures of TB containing ampicillin at concentrations ranging from 0 to 1200 μg/mL, increasing in increments of either 100 μg/mL or 200 μg/mL, depending on the experiment. In all tests, ultimate cell culture densities and the quantities of plasmid DNA extracted from 1.5 mL minipreps were similar, regardless of ampicillin concentration (Figure 4A and 4B). Somewhat surprisingly, cultures containing no added ampicillin generated as much DNA as those that contained the antibiotic. These results indicate that increasing selective pressure by supplementing with higher amounts of ampicillin does not enhance DNA yields.
Fig. 4.
Elevated concentrations of ampicillin do not enhance (A) cell growth or (B) plasmid DNA yield or quality. Top10 cells containing pET15b-rKGA were harvested from LB plates containing 100 μg/mL ampicillin and shaken overnight at 37°C in TB broth containing the indicated amounts of ampicillin. Averages and standard deviations from four cultures are depicted in (A).
In conclusion, we have analyzed several commonly used E. coli cell culture broths and determined that TB produced the highest yields of low copy number plasmid DNA from minipreps. This result is consistent with some previous studies of protein expression in E. coli cells that noted improved yields of protein when TB was used (Li et al., 1990; Nakamura et al., 1999; Losen et al., 2004; David et al., 2015). The superiority of TB for DNA purification in the current study was linked to its high content of yeast extract and was not dependent upon glycerol, a unique energy source in TB that has been described as an important carbon source in this media (Tartoff and Hobbs, 1987; da Silva et al., 2009; Lessard, 2013). We observed that simply adding an equivalent amount of yeast extract (24 g/L) to LB resulted in DNA yields similar to those seen with TB. This result demonstrates that complex broths such as SOC, TB, and more exotic alternatives such as H15 which contain supplemental buffers, salts and additional carbon sources (Tartoff and Hobbs, 1987; Duttweiler et al., 1998; Lessard, 2013) are not needed to generate high plasmid DNA yields.
Recovery of the single copy number plasmid pBeloBAC11 was also enhanced using high yeast extract broths. Yields of these plasmids were quite low, typically producing ~ 200 ng total DNA from 1.5 mL minipreps, versus ≥ 5000 ng with the low copy number plasmids. However, these quantities were more than sufficient for analysis by gel electrophoresis and are likely to be sufficient for many downstream applications. Finally, we note that yields of the low copy number plasmids could not be improved by increasing the concentration of antibiotic in the growth media, i.e., by exerting greater selective pressure for retention of the plasmids by the cells.
Highlights.
TB Broth produced the greatest yields of low copy number plasmid DNA from minipreps
Yeast extract, not glycerol carbon source, was key to superior results seen with TB
Increasing yeast extract in the most commonly used broth, LB, made it equal to TB
Adding more ampicillin to enhance selection for plasmids did not improve DNA yields
Acknowledgments
The authors would like to thank Joseph Wortman for help with the project. LKL was supported by a grant from the National Institutes of Health (Grant 1R15GM09904901).
Footnotes
Conflict of interest
The authors declare that they have no conflict of interest.
Contributors
WNW and KDS performed experiments assessing yields of low copy plasmids using different broths and antibiotic concentrations, JAR and LKL performed tests involving single copy plasmids, and LKL wrote and edited the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Cheong DE, Choi JH, Song JJ, Kim GJ. Construction of non-invasively constitutive expression vectors using a metagenome-derived promoter for soluble expression of proteins. Bioprocess Biosyst Eng. 2013;36:667–76. doi: 10.1007/s00449-013-0890-x. [DOI] [PubMed] [Google Scholar]
- Christianson TW, Sikorskim RS, Dante M, Shero SH, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- da Silva GP, Mack M, Contiero J. Glycerol: a promising and abundant carbon source for industrial microbiology. Biotechnol Adv. 2009;27:30–39. doi: 10.1016/j.biotechadv.2008.07.006. [DOI] [PubMed] [Google Scholar]
- David B, Torgeman A, Barnea A, Zichel R. Expression, purification and characterization of the receptor-binding domain of botulinum neurotoxin serotype B as a vaccine candidate. Protein Expr Purif. 2015;110:122–129. doi: 10.1016/j.pep.2015.02.008. [DOI] [PubMed] [Google Scholar]
- Dersch P, Fsihi H, Bremer E. Low-copy-number T7 vectors for selective gene expression and efficient protein overproduction in Escherichia coli. FEMS Microbiol Lett. 1994;123:19–26. doi: 10.1111/j.1574-6968.1994.tb07195.x. [DOI] [PubMed] [Google Scholar]
- Duttweiler HM, Gross DS, Saeki MK, Takahashi Y. Bacterial growth medium that significantly increases the yield of recombinant plasmid. BioTechniques. 1998;24:438–444. doi: 10.2144/98243st03. [DOI] [PubMed] [Google Scholar]
- Elbing K, Brent R. Curr Protoc Mol Biol. 2002:1.0.1–1.3.6. [Google Scholar]
- Farrell SO, Taylor LE. Experiments in Biochemistry: A Hands-on Approach. 2. Thomson-Brooks/Cole Laboratory Press; Belmont, CA: 2005. [Google Scholar]
- Feinbaum R. Curr Protoc Mol Biol. 2002:1.5.1–1.5.17. doi: 10.1002/0471142727.mb0105s41. [DOI] [PubMed] [Google Scholar]
- Green MR, Sambrook J. Molecular Cloning: a Laboratory Manual. 4. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2012. [Google Scholar]
- Guan L, Liu Q, Li C, Zhang Y. Development of a Fur-dependent and tightly regulated expression system in Escherichia coli for toxic protein synthesis. BMC Biotechnol. 2013;13:25. doi: 10.1186/1472-6750-13-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard JC. Growth media for E. coli. Methods Enzymol. 2013;533:181–189. doi: 10.1016/B978-0-12-420067-8.00011-8. [DOI] [PubMed] [Google Scholar]
- Li XL, Robbins JW, Taylor KB. The production of recombinant beta-galactosidase in Escherichia coli in yeast extract-enriched medium. J Ind Microbiol. 1990;5:85–93. doi: 10.1007/BF01573857. [DOI] [PubMed] [Google Scholar]
- Losen M, Frölich B, Pohl M, Büchs J. Effect of oxygen limitation and medium composition on Escherichia coli fermentation in shake-flask cultures. Biotechnol Prog. 2004;20:1062–1068. doi: 10.1021/bp034282t. [DOI] [PubMed] [Google Scholar]
- Mardanov AV, Strakhova TS, Smagin VA, Ravin NV. Tightly regulated, high-level expression from controlled copy number vectors based on the replicon of temperate phage N15. Gene. 2007;395:15–21. doi: 10.1016/j.gene.2006.12.036. [DOI] [PubMed] [Google Scholar]
- Nakamura M, Saeki K, Takahashi Y. Hyperproduction of recombinant ferredoxins in Escherichia coli by coexpression of the ORF1-ORF2-iscS-iscU-iscA-hscB-hscA-fdx-ORF3 gene cluster. J Biochem. 1999;126:10–18. doi: 10.1093/oxfordjournals.jbchem.a022409. [DOI] [PubMed] [Google Scholar]
- Nordström K, Dasgupta S. Copy-number control of the Escherichia coli chromosome: a plasmidologist’s view. EMBO Rep. 2006;7:484–489. doi: 10.1038/sj.embor.7400681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers D. Timing, self-control and a sense of direction are the secrets of multicopy plasmid stability. Mol Microbiol. 1998;29:1137–1145. doi: 10.1046/j.1365-2958.1998.01012.x. [DOI] [PubMed] [Google Scholar]
- Tartoff KD, Hobbs CA. Improved media for growing plasmid and cosmid clones. Bethesda Res Lab Focus. 1987;9:12. [Google Scholar]
- Tillett D, Neilan BA. Small-scale preparation of the single-copy bacterial artificial chromosome vector pBeloBAC11. Biotechniques. 1998;24:568–572. doi: 10.2144/98244bm10. [DOI] [PubMed] [Google Scholar]