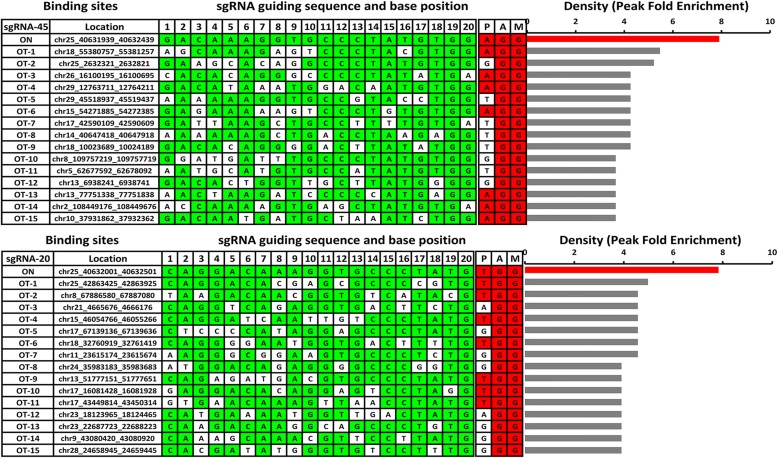
Fig. 3.
Off-target sites of sgRNA 45 and 20 with the top 15 ChIP-seq binding densities. All of the off-target sites were computationally identified with 20 bp sequences. These sites ended with PAM and aligned most effectively with the sgRNA guiding sequences in each peak. OT indicates off-target; all of the off-target sites in the same group were ranked according to the ChIP-seq binding densities (peak fold enrichments) as illustrated in the bar graphs on the right. At the off-target sites, bases matching the sgRNA guiding sequence and the PAM sequence are highlighted in green and red, respectively. Similar results for sgRNA 2 and 20 are available in Additional file 3: Figure S3