FIG 2.
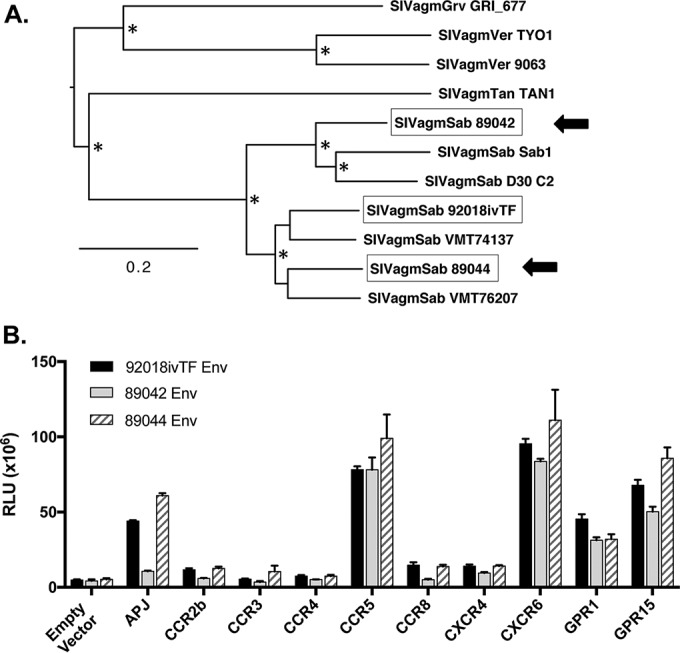
Divergent field isolate SIVagmSab Envs efficiently use alternative coreceptors in vitro. SIVagmSab Envs were amplified from the plasma of two wild-infected sabaeus monkeys (animals 89042 and 89044). (A) Maximum likelihood phylogenetic tree showing SIVagmSab89042 and SIVagmSab89044 env genes as well as SIVagmSab92018ivTF and other previously described SIV env nucleotide sequences. Envs used in this study are boxed, and the two env genes cloned here from wild, naturally infected animals are indicated by arrows. Bootstrap values of >70% are indicated by an asterisk. The bar indicates 0.2 nucleotide substitutions per site. (B) 293T cells were transfected with each sabaeus candidate coreceptor in conjunction with sabaeus CD4-31. Cells were then infected with luciferase reporter pseudotypes containing field isolate SIVagmSab Envs or SIVagmSab92018ivTF Env. Cells were lysed and the luciferase content was measured 72 h later. Infections were carried out in triplicate, and data (means ± standard deviations) are representative of results from 3 replicate experiments.
