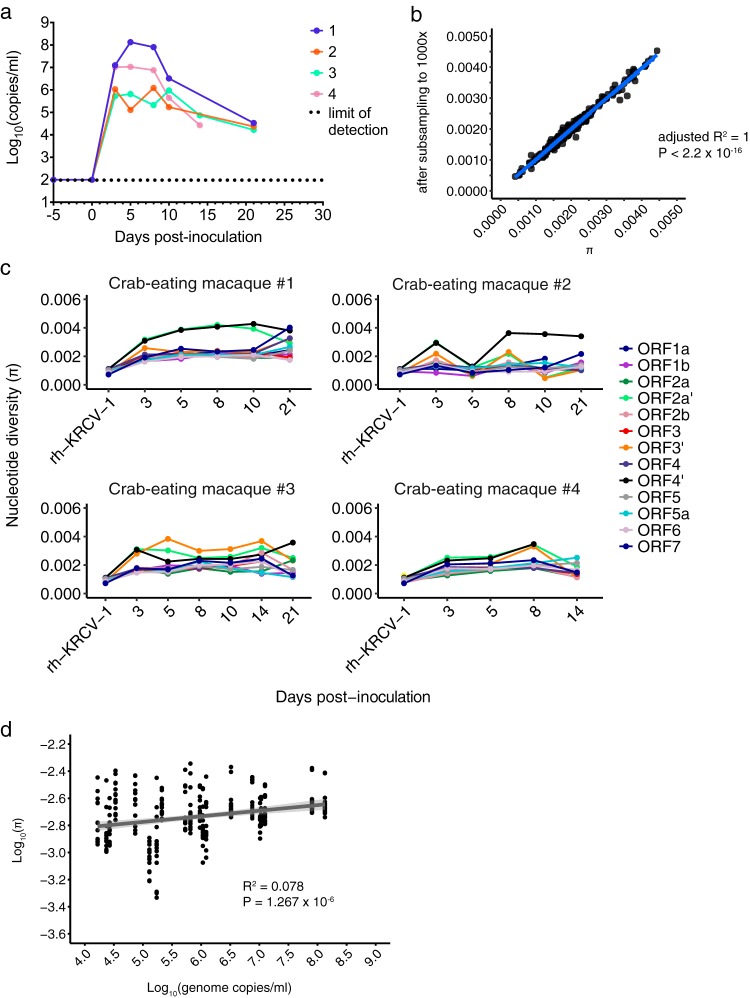
FIG 3.
KRCV-1 selection in infected macaques is weak. (a) Log10 viral load in viral RNA copies/ml plasma from each crab-eating macaque infected with KRCV-1. Viral loads were quantified using qRT-PCR. The numbers 1 to 4 indicate the crab-eating macaque animal number. Data taken from reference 17 were replotted here. (b) π was calculated using either the original assemblies or assemblies subsampled to produce even coverage of 1,000× across the entire genome. For each ORF, animal, and time point, the corresponding π value calculated from the original assemblies is plotted on the x axis versus the π value for the same sample calculated from the subsampled assembly on the y axis. The results obtained were very similar, suggesting that uneven genome coverage likely did not impact diversity estimates. (c) KRCV-1 genome nucleotide diversity (π) was estimated for each ORF, day of infection, and infected crab-eating macaque (numbered 1 to 4). (d) The log-transformed π values for every ORF are plotted against the log-transformed viral load values. Regression analysis was performed in R.