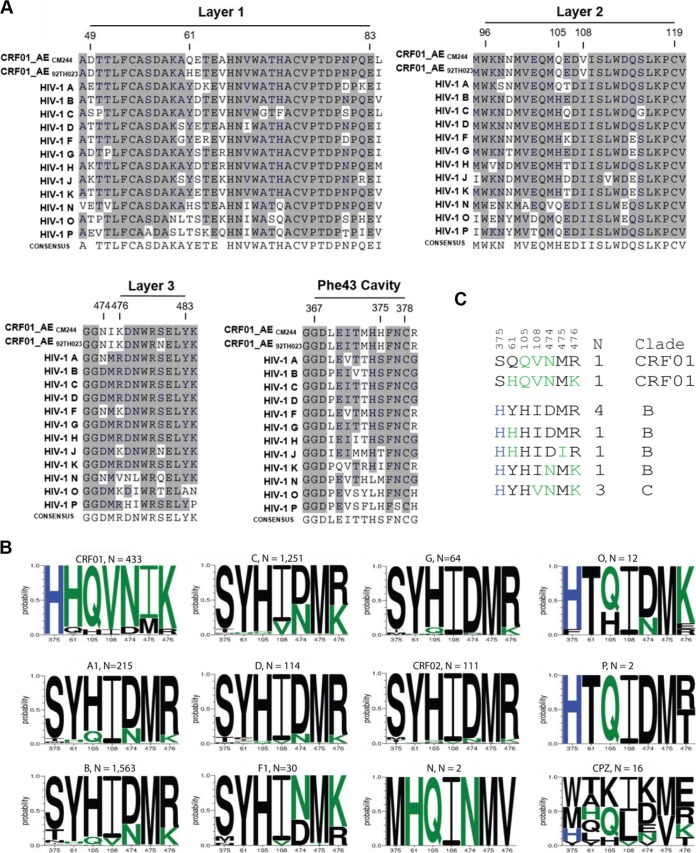
FIG 1.
Phe43 cavity and inner-domain layer HIV-1 Env sequences. (A) Primary sequence alignment of gp120 residues form the Phe43 cavity, layer 1, layer 2, andlayer 3 from a single sequence from each clade: HIV-1 A (accession number ABB29387.1), HIV-1 B (accession number K03455), HIV-1 C (AAB36507.1), HIV-1 D (P04581.1), HIV-1 F (ACR27173.1), HIV-1 G (ACO91925.1), HIV-1 H (AAF18394.1), HIV-1 J (ABR20452.1), HIV-1 K (CAB59009.1), HIV-1 N (AAT08775.1), HIV-1 O (AAA99883.1), HIV-1 P (GQ328744), and two studied viruses, CRF01_AE CM244 (AY713425) and CRF01_AE 92TH023. Residue numbering is based on that of the HXBc2 strain of HIV-1. The shading highlights residues that are similar to those of CRF01_AE from the other subtype from group M. (B) A Logo depiction of the frequency of each amino acid in the positions that we originally noted by eye, based on the data shown in panel A, to be correlated with 375 H and 375 S/T. The height of the letter indicates its frequency. CRF01_AE has >99% 375 H, and the amino acids that are shown to be enriched among CRF01_AE sequences, therefore also associated with 375 H, are shown in green. Black indicates amino acids that are favored in other clades or are rare variants. Logos for sequences from the following subtypes and groups are shown: HIV-1 M group major subtypes A1, B, C, D, F1, and G; M group CRFs 01 and 02; HIV-1 groups N, O, and P; and Envs isolated from chimpanzees (CPZ). Subtype and groups are noted above each Logo, followed by their count. The 2016 Los Alamos database curated Env alignment was used as the basis for this figure, which contains 4,632 HIV-1 and related SIVcpz sequences. (C) Alignment of the positions of interest in the inner domain, ordered as described for panel B. The two instances of an H375S being found in CRF01_AE are indicated, and the 10 naturally occurring sequences from other M group major subtypes with the S375H mutation are also shown. As in panel B, CRF01_AE common amino acids are shown in green, the positions are indicated above the alignment, and the number of times the pattern was found (N) and the clade in which the variant arose are noted on the right.