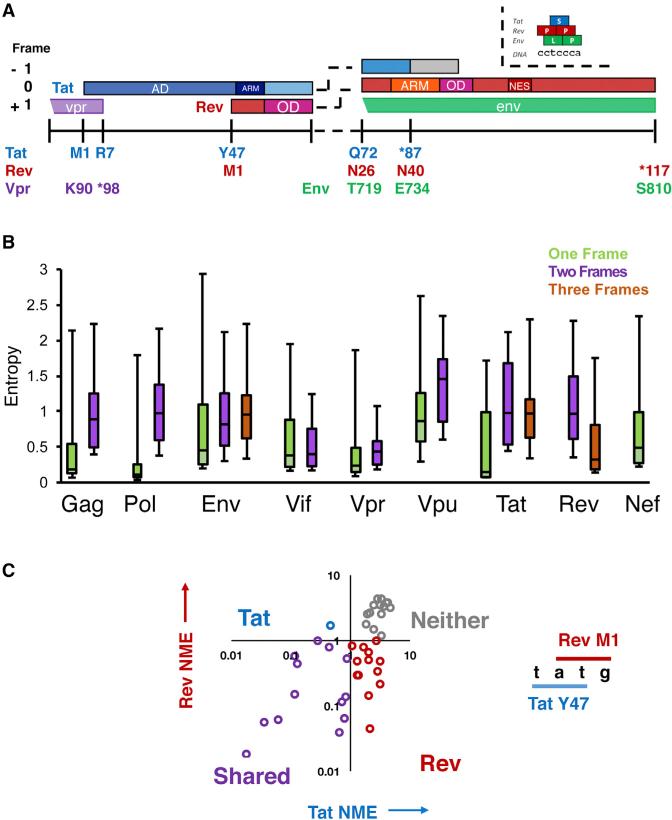
Figure 1. Organization and Conservation of HIV-1 Overlaps.
(A) Layout of the Genetic Organization of the tat/rev overlap in HIV-1. ARM, arginine rich motif/nuclear localization sequence; OD, oligomerization domain; NES, nuclear export sequence. In HIV-1 NL4-3 Tat is 86 residues, although many patient genes are 101 residues (gray box).
(B) Individual gene entropy analysis for overlapped and single-frame regions in the HIV-1 genome (see Figure S2A). Entropy values were computed at the protein level for each frame and Shannon entropy values for alignments of HIV-1 patient sequences are shown. Median, range, and interquartile range (IQR) are shown in the box and whiskers plot. A score of 0 indicates absolute conservation and a score of 3 indicates near-absolute degeneracy.
(C) Categorization of sites by normalized mean entropy (NME) in the tat/rev overlap. Residues are grouped into pairs that share two nucleotides, and their NME plotted accordingly (Tat NME, Rev NME). Quadrants are labeled to indicate which genes are conserved in that region.
See also Figure S1.