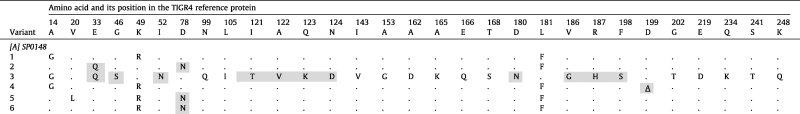
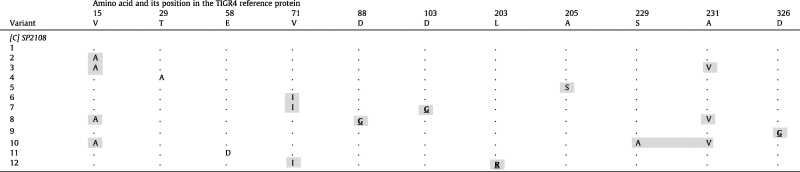
Table 3.
Amino acid variations in three S. pneumoniae protein vaccine candidates [A] SP0148, [2] SP1912 and [C] SP2108, among a global dataset of serotype 1 pneumococci. The tables show the amino acid residues at which substitutions have been identified when using the corresponding protein sequences from Streptococcus pneumoniae TIGR4 as a reference. ‘.’ Indicates that a residue is identical to the TIGR4 reference sequence. Residues highlighted in grey indicate regions where the amino acid variation is predicted to result in a change to the antigenicity profile of the protein. Underlined residues indicate that the amino acid variation is predicted to be deleterious to the protein. Amino acid numbering starts at the translation site (the methionine) of the TIGR4 reference sequences.
 |
 |
 |
