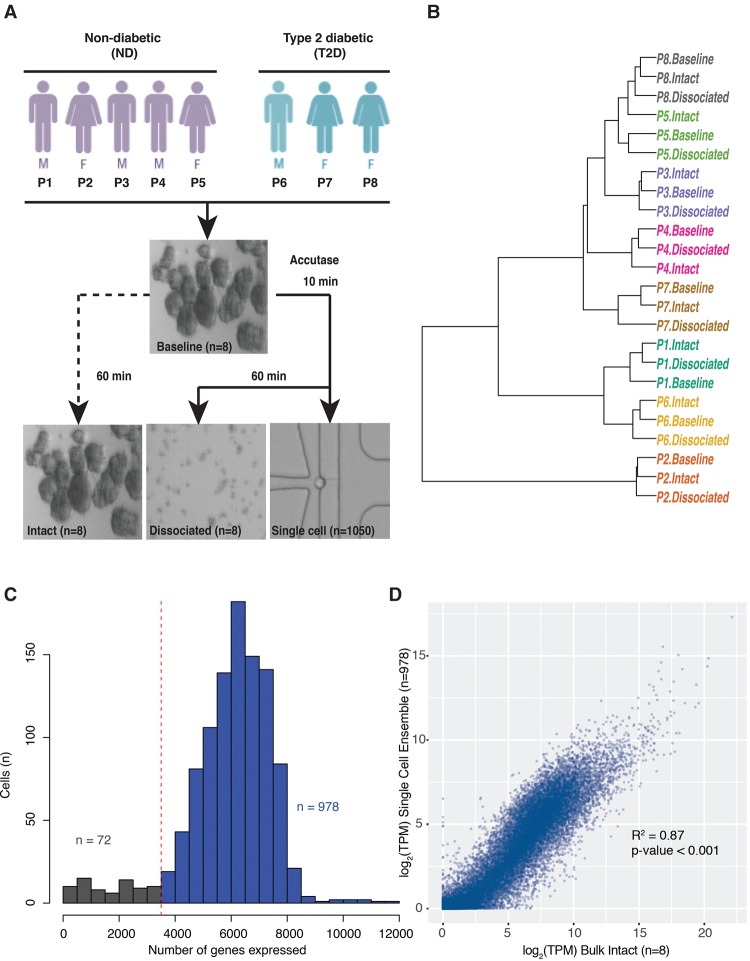
Figure 1.
Single-cell transcriptomes reflect those of paired intact islets. (A) Schematic of experimental workflow. Islets from each donor sample (n = 8 individuals) were dissociated using Accutase, and single-cell transcriptomes were synthesized from 1050 cells captured using 11 Fluidigm C1 chips. In parallel, “bulk” RNA-seq libraries were prepared from remaining dissociated single cells (dissociated) and from intact islets either flash frozen (baseline) or incubated/processed (intact). (B) Unsupervised hierarchical clustering of baseline, intact, and dissociated islet transcriptomes demonstrates clustering by person and not by processing/experimental condition. (C) Histogram demonstrating the number of genes detected in each single cell. Cells expressing less than 3500 genes (n = 72) were removed from downstream analyses. (D) Scatter plot comparing intact islet bulk RNA-seq (n = 8) and ensemble single-cell RNA-seq (n = 978) data demonstrates high correlation. (R2) Pearson's R-squared; (TPM) transcripts per million; (P) person.