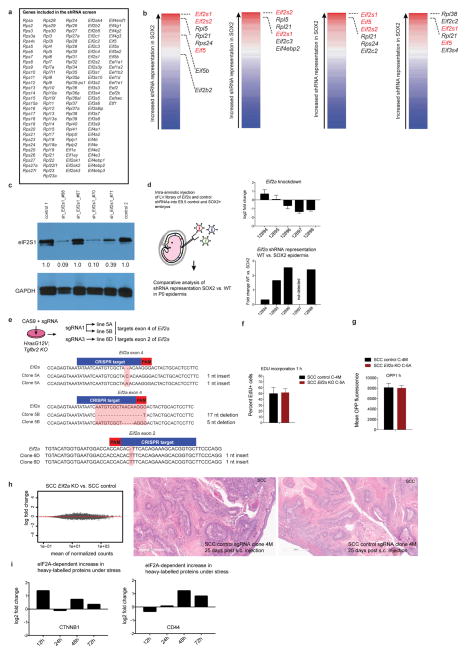
Extended Data Figure 7. An shRNA screen reveals regulatory nodes in premalignancy.
a, The 138 ribosomal genes and initiation factors targeted in our shRNA screen. b, Overview of the shRNA screen in wild-type versus premalignant SOX2-expressing epidermis from P0 mice. The RIGER algorithm27 was used with the following methods and metrics to convert hairpins to genes and to rank top hits. From left to the right: weighted sum, signal to noise (median); second best rank, signal to noise (median); weighted sum, signal to noise; weighted sum, fold change. c, Western blot shows knockdown efficiency of shRNAs targeting the top hit in our screen, Eif2s1. Note that the knockdown efficiency correlates well with the degree of shRNA depletion in the screen. d, Intra-amniotic injection of lentivirus library of Eif2a and 35 control shRNAs. This sub-library was injected into E9.5 embryos and representation of shRNAs was quantified in wild-type and SOX2 P0 skin. Top, knockdown efficiency; bottom, relative representation of Eif2a shRNAs (normalized against the control shRNA library) in wild-type versus SOX2 epidermis. e, DNA sequence of Eif2a knockout clonal cell lines used in our study. PAM region is highlighted in red, CRISPR target region in blue. f, g, Proliferation rates and overall protein synthesis rates are unchanged in Eif2a knockout cells under serum-rich conditions in vitro. SCC control and Eif2a knockout cells were quantified for EdU and OPP incorporation 1 h after administration. Data are mean ± s.d. of 4 independent experiments. h, The translational landscape is unchanged in Eif2a knockout cells under serum-rich conditions in vitro. SCC control and Eif2a knockout cells subjected to ribosome profiling and reads within the main ORFs were quantified and tested for differential expression using DESeq2. As opposed to 5′ UTR translation, no significant differences (adjusted P < 0.1 DESeq2) in ORF translation were found between SCC control and SCC Eif2a knockout cells (n = 2 SCC control, n =2 SCC Eif2a knockout). Right panels show two representative H&E sections of squamous cell carcinomas formed 25 days after subcutaneous injection of SCC cells. i, eIF2A-dependent changes of heavy-labelled peptides of CTNNB1 and CD44 under stress in pulsed SILAC. SCC control and SCC Eif2a knockout cells were grown in light-labelled medium and switched to heavy-labelled medium supplemented with 5 μM arsenite. Graphs show the relative difference between SCC control and SCC Eif2a knockout cells during the pulsed SILAC time course.