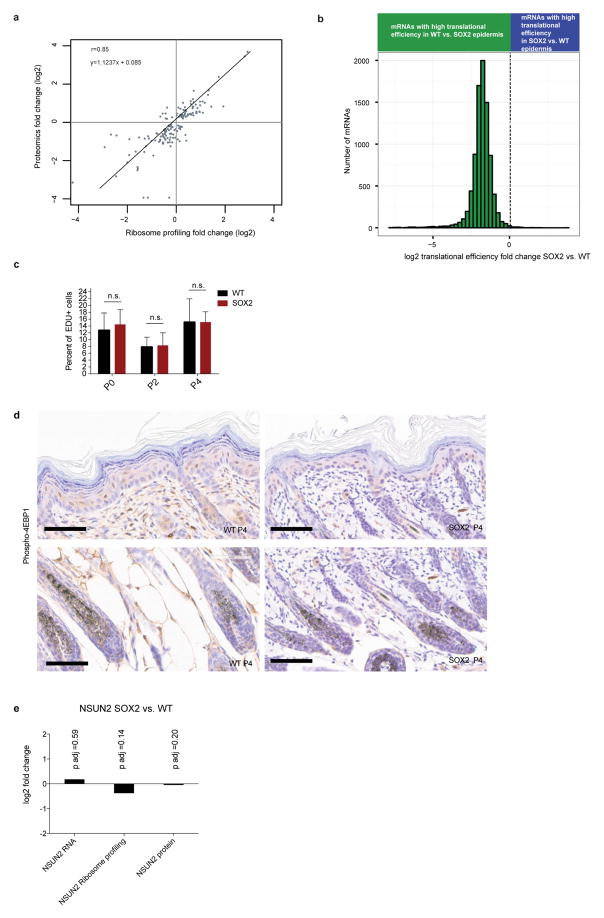
Extended Data Figure 2. Global translational efficiency is decreased upon SOX2 expression in keratinocytes in vitro.
a, Ribosome profiling data correlates with proteomics data. At a stage when proliferation and morphology were similar, freshly isolated, basally enriched keratinocytes from wild-type and SOX2-expressing P4 skins were subjected to in vivo ribosome profiling and to a label-free proteomics strategy. Plotted are proteomics fold changes compared to ribosome profiling fold changes. Comparisons are made in SOX2 versus wild-type samples for significantly changed proteins (false discovery rate (FDR) <0.05). b, Translational efficiency is markedly reduced in SOX2-expressing premalignant keratinocytes in vitro. Keratinocytes were isolated from R26-Sox2-IRES-eGFPfl/+ (WT) or K14-cre; R26-Sox2-IRES-eGFPfl/+ (SOX2+) P0 animals and cultured in vitro. Quantified were genes with more than 256 reads in RNA-seq data. Histogram shows distribution of differential translational efficiency (TE =RPKMribosome profiling/RPKMRNA-seq). Data are shown from 4 independent ribosome profiling experiments and 2 independent RNA-seq experiments. c, Wild-type and SOX2 keratinocytes have similar proliferation rates in vivo. Basal epidermal EdU incorporation in P0, P2 and P4 mice (n = 442/496, 385/449 and 903/841 cells from duplicate wild-type/SOX2 animals) 1 h after injection. Data are mean ±s.d. d, Phospho-4E-BP1 immunohistochemistry in the epidermis shows no difference in levels upon SOX2 induction. e, NSUN2 transcript, translation, and protein levels in SOX2 versus WT P4 epidermis.