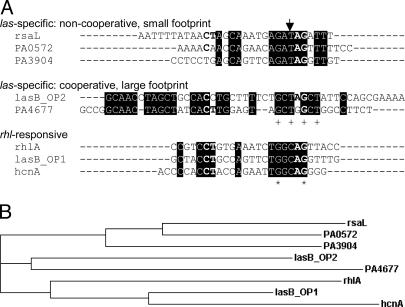
Fig. 7.
Sequence analysis. (A) Alignment of binding sequences in quorum-controlled promoters performed with clustalw (www.ebi.ac.uk/clustalw). Depicted are the sequences protected from DNase I digestion as determined from Figs. 1D and 6. The sequence of the rhlA promoter that did not bind LasR but presumably binds RhlR was extrapolated based on the location of a predicted las–rhl box sequence and the footprinting pattern that LasR exhibits when it binds to promoters with no cooperativity. Elements of the CT-[N]12-AG motif previously thought to be absolutely conserved (12, 31) are shown in bold. The arrow designates a base position potentially conferring las vs. rhl sequence specificity. + indicates base positions identical in all five las-specific sequences. Asterisk indicates base positions identical in all eight sequences shown. (B) Phylogram (hierarchical clustering) of the sequences shown in A.