Figure 4.
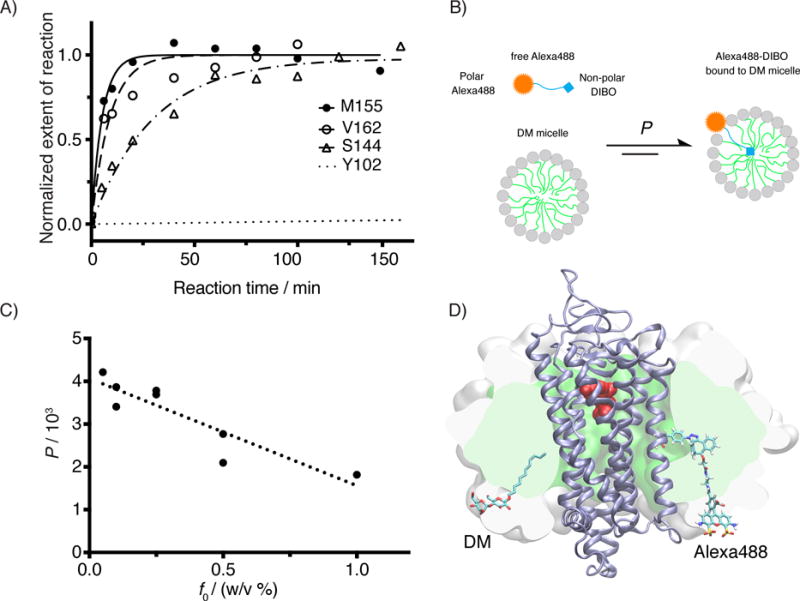
Kinetic study for the SpAAC reaction between azF-Rho and Alexa488-DIBO. A) Reaction time course of four azF-Rho variant with Alexa488-DIBO (10 μM): M155 and V162 (TM4), S144 (IC2) and Y102 (EC2). The data points were fitted using pseudo first-order kinetic model. The calculated k2 is (3.2 ± 0.4)×102 M−1 s−1 for M155azF-Rho, and (1.9 ± 0.5) ×102 M−1 s−1 for V162azF-Rho. The kinetic study for S144azF-Rho (k2 = 62 M−1 s−1) has been described earlier[10c] and included in the plot for comparison. The curve for Y102azF-Rho was simulated based on the k2 (= 0.27 M−1 s−1) estimated from single time point labeling stoichiometry data.[10c] B) Partition equilibrium of Alexa488-DIBO between water and DM micelle. C) Partition coefficients (water to micelle) from ultrafiltration experiment for various concentrations of DM. The observed partition coefficients (P) were plotted as against the mass concentration of DM (f0). We found empirically a linear correlation between P and f0 (R2 = 0.81). D) Atomic model of Rho (PDB:1GZM) embedded in a micelle (150 DM molecules). The SpAAC conjugated V162azF-DIBO-Alexa488 is shown as sticks including hydrogens. For size comparison, one of the 150 DM molecules is shown in sticks but without hydrogens. The protein backbone is shown as a cartoon (ice blue). 11-cis-retinal is shown in spheres (red). The hydrophobic core of the micelle is rendered as a green surface sliced by the image plane just behind the dye. The white surface shows the extension of the head groups of the micelle sliced in the same plane. The model of Alexa488-DIBO was generated with Schrödinger’s small-molecule drug discovery suite, and the model of DM micelle was constructed by charmm-gui micelle builder.[53] A short molecular dynamics simulation (1.3 ns) was performed with namd2.10[54] to relax the DM micelle around the receptor.
