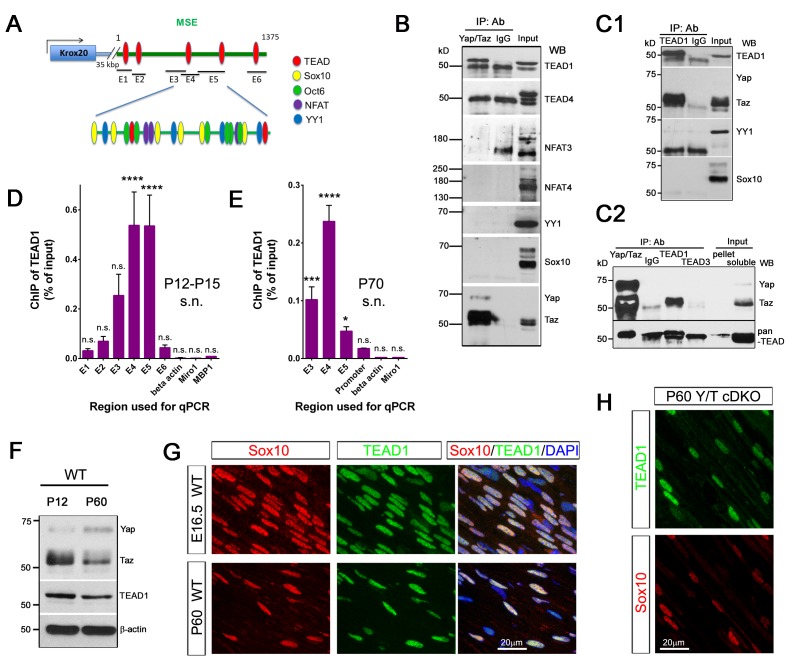
Figure 8. YAP/TAZ-TEAD1 regulate Krox20 transcription during developmental myelination and adult myelin maintenance.
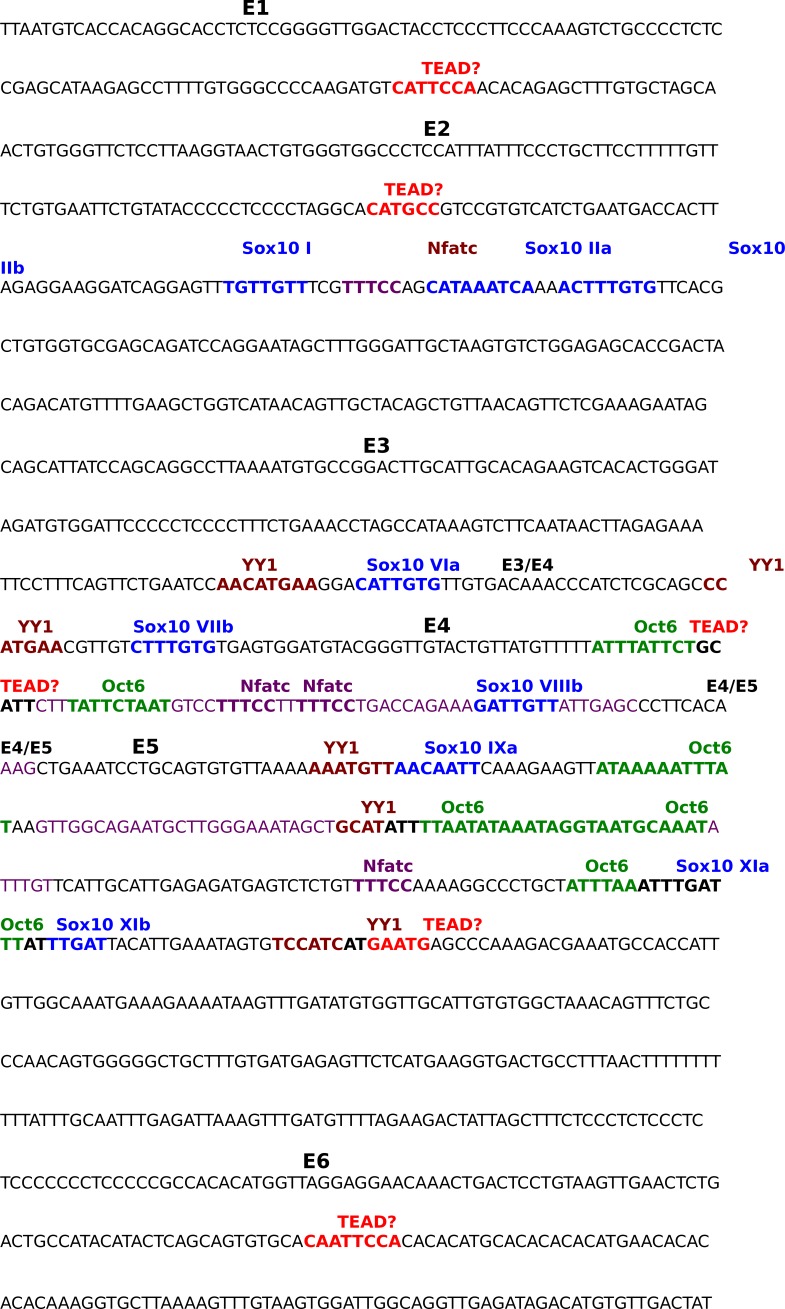
(A) Schematic showing potential TEAD binding sites in the Krox20 MSE, in relation to published binding sites for known MSE-binding transcription factors. The regions of the MSE covered by primer sets E1-E6 used for ChIP-qPCR are indicated. (B, C1, C2) Immunoprecipitation of P12-P15 WT sciatic nerve lysates with anti-YAP/TAZ (B), anti-TEAD1 (C1), anti-YAP/TAZ, anti-TEAD1 and anti-TEAD3 (C2), plus control Rb IgG (B, C1, C2). Immunoprecipitates were blotted with the indicated antibodies. In (C2), upper blot was stripped and reprobed with anti-pan-TEAD (lower blot): the band in the Rb IgG lane, migrating similarly to TEAD proteins, is non-specific. n = 3 (B) or n = 2 (C1, C2) experiments. (D, E) ChIP-qPCR of P12-P15 sciatic nerves (D) or P70 sciatic nerves (E), using anti-TEAD1 antibody or control Rb IgG. Graphs show the amount of qPCR product obtained with the indicated MSE-specific primer sets or negative control primer sets after TEAD1 ChIP. Amounts are given as % of qPCR product obtained with the same primers using input chromatin. Amounts obtained with the same primer sets after control Rb IgG ChIP were all less than 0.003% of input, and are not shown. (D) n = 5 (E1, E2, E4, E6), n = 4 ((E5), n = 3 (β-actin), n = 2 (E3) independent ChIP assays. (E) n = 2 independent ChIP assays. P-values are given for TEAD1 ChIP versus Rb IgG ChIP for each primer set. ****p<0.0001, ***p=0.0001, *p=0.0498, n.s. = non-significant, 2- way ANOVA with Sidak’s multiple comparison test. Bar represents mean ± SEM. (F) Western blotting of sciatic nerve lysates from WT of P12 or P60 mice, showing persistent expression of TEAD1 and YAP/TAZ. (G) Immunostaining of longitudinal sections of WT sciatic nerves, showing that TEAD1 (green) is present in all nuclei (marked by Sox10, red) of proliferating and differentiated SCs. (H) Longitudinal nerve sections of P60 Yap/Taz cDKO, showing that TEAD1 (green) is present in all cDKO SC nuclei (marked by Sox10, red). The following figure supplements are available for Figure 8.