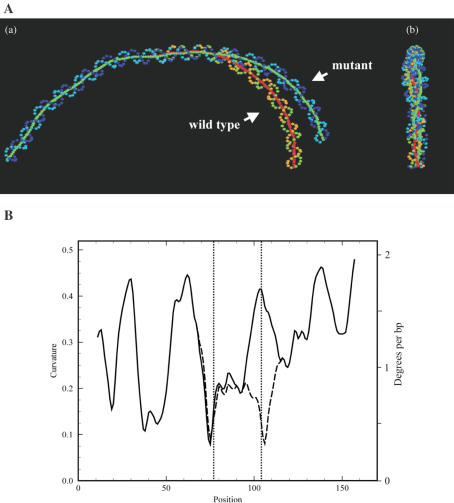
Figure 5.
Juxtaposition of predicted DNA curvature for wild-type and modified sequences parC. The wild type and modified sequences were applied to the software CURVATURE (80) with the specified parameters (52,81). Both fragments are 167 bp long. The modifications were made at positions 77, 79, 104 and 105 relative to this fragment length (corresponding to sites 192, 194, 219 and 220 in the numbering frame of Figure 2). (A) Two-dimensional representation of the DNA paths. Two projections of the DNA paths are presented on a plane of the maximal observed curvature (a) and on a reciprocal plane (b). To display the degree of curvature of a fragment, the calculated coordinates of the phosphates and base pair centers are projected on that plane, for which the mean square distance from all the points is minimal. The line connecting the base pair centers presents the helical axes of the original (wild type: green continued by red) and modified fragment (mutant: green). The phosphates are shown in light and dark blue (mutant) switching to light brown and green for the wild type. The second projection (right) shows the planarity of the fragments. The two base pair exchanges at 219 and 220 bp [resp. 104 and 105 in (B)] result in a curvature reduction at this site of 17 degrees. (B) DNA curvature maps versus sequence with window-size of 21 bp. The overall DNA shapes are characterized curvatures of a sliding arc of 21 bp of length approximating to the paths of the axes of the given DNA fragments. The curvature map (given in ‘cu units’ on the left axis and ‘degrees per bp’ on the right axis) for wild-type parC is plotted as a solid line and for the mutant by a dashed line. The two vertical dotted lines indicate the positions of the base pair modifications.