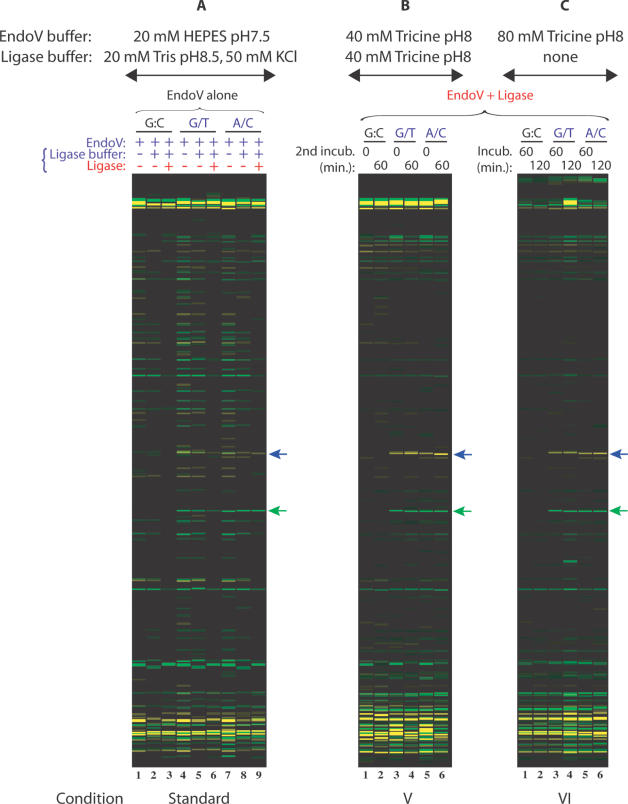
Figure 3.
Comparison of a single-step assay, a standard two-step assay, and a combined two-step EndoV/ligase assay in Tricine buffer. Universal PCR primers (VicUniEV1F/NedUniEV2R), along with gene-specific primers F173 and R174, were used to amplify 350 bp fragments of p53 exon 8 carrying R273H (G→A) mutation. VIC- and NED-labeled mutation cleavage products are 158 nt (green arrow) and 195 nt (blue arrow) in size. The ‘split label, denaturation, renaturation’ method was used to generate heteroduplexes of either top strand wild-type:bottom strand mutant (G/T mismatch) or top strand mutant:bottom strand wild-type (A/C mismatch). Wild-type PCR product mixtures were used as controls (G:C match). EndoV/ligase assay conditions tested are briefly indicated on top of the figure and detailed in Table 2. (A) Standard EndoV/ligase procedure; (B) Condition V is a two-step reaction combining both enzymes in the first incubation step [EndoV + ligase], followed by a second incubation in ligase buffer; (C) Condition VI is a single-step reaction with both EndoV and ligase. Reaction mixtures were electrophoresed on the ABI 3730 fluorescence-based capillary electrophoresis instrument. Data were analyzed using Gene Mapper fragment analysis software.