FIGURE 1.
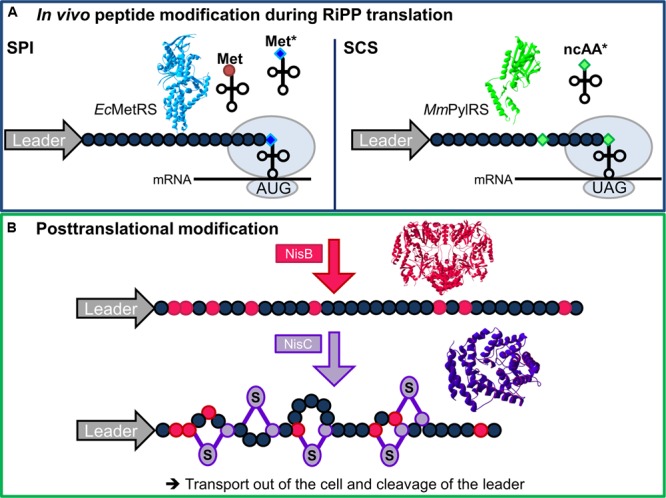
Chemical diversification of AMPs by ncAAs. (A) In vivo prepeptide modification by ncAAs during RiPP translation (core residues in dark blue circles). Incorporation of ncAAs (∗) can be achieved by two methods: Selective pressure incorporation (SPI, left) allows installation of isostructural variants of canonical amino acids, here as an example Met variants (blue diamonds) charged onto tRNAMet by the endogenous E. coli methionyl-tRNA synthetase (MetRS, light blue, PDB ID 1PG2; Crepin et al., 2003). The second method, stop codon suppression (SCS, right), requires co-expression of an orthogonal pair. A suppressor tRNA (here recognizing the amber stop codon UAG) is charged with the target ncAA (green diamonds) by its corresponding aminoacyl-tRNA synthetase (e.g., PylRS from Methanosarcina mazeii, depicted in green, PDB ID 2Q7H; Kavran et al., 2007). (B) Posttranslational AMP modifications with the model lantibiotic nisin as chosen example. First, dehydration of certain prepeptide serine and threonine residues catalyzed by the dehydratase NisB (both in magenta, PDB ID 4WD9; Ortega et al., 2015) yields dehydroalanines and dehydrobutyrines, respectively. Subsequent cyclization with Cys residues by the cyclase NisC (both purple, PDB ID 2G0D; Li et al., 2006) affords the characteristic (methyl-)lantionine rings. The depicted elements are not true to scale. 3D structures of proteins rendered with Swiss PDB viewer version 4.1.0.
