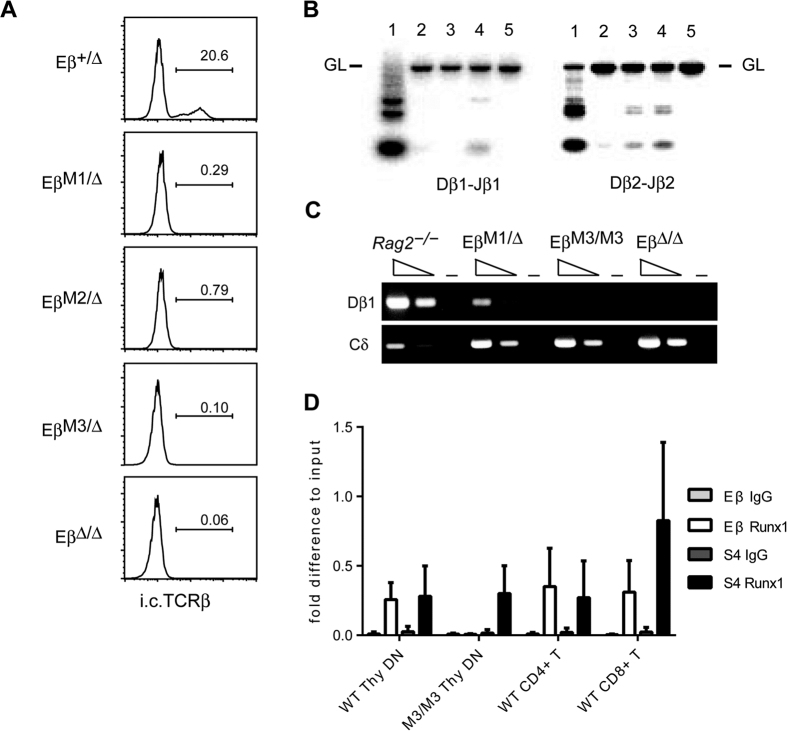
Figure 2. Effect of Runx site mutations on DJ recombination and germline transcription.
(A) Histograms showing the expression of intracellular TCRβ (i.c.TCRβ) in a CD25+ CD44− DN3 subset from mice with the indicated genotype. (B) Semi-quantitative DNA-PCR analyses for analyzing Dβ to Jβ rearrangement in CD25+ CD44− DN3 subsets from Eβ+/Δ(lane 1), EβΔ/Δ (lane 2), EβM1/Δ (lane 3), EβM2/Δ (lane 4) and EβM3/Δ (lane 5) mice. The bar indicates the position corresponding to the germline (GL) configuration. (C) The germline transcript of the Dβ1 region in CD4−CD8− DN cells isolated from Rag2−/−, EβM1/Δ, EβM3/M3 and EβΔ/Δ mice are shown. The germline transcript of Cδ region was used as control. RNA without reverse-transcriptase reaction is shown in lanes indicated as (−). (D) Chromatin immunoprecipitation assay (ChIP) to detect Runx1 binding to Eβ and Cd4 silencer (S4). CD4−CD8− DN cells isolated from wild-type and EβM3/M3 mice were used to prepare chromatin DNA. Chromatin DNA was immunoprecipitated with the IgG control or anti-Runx1 antibody and was used as the template for qPCR amplification. The Cd4 silencer region as well as peripheral CD4+ and CD8+ T cells (WT) were used as controls for Runx1 binding. Combined data from three independent ChIP experiments is shown.