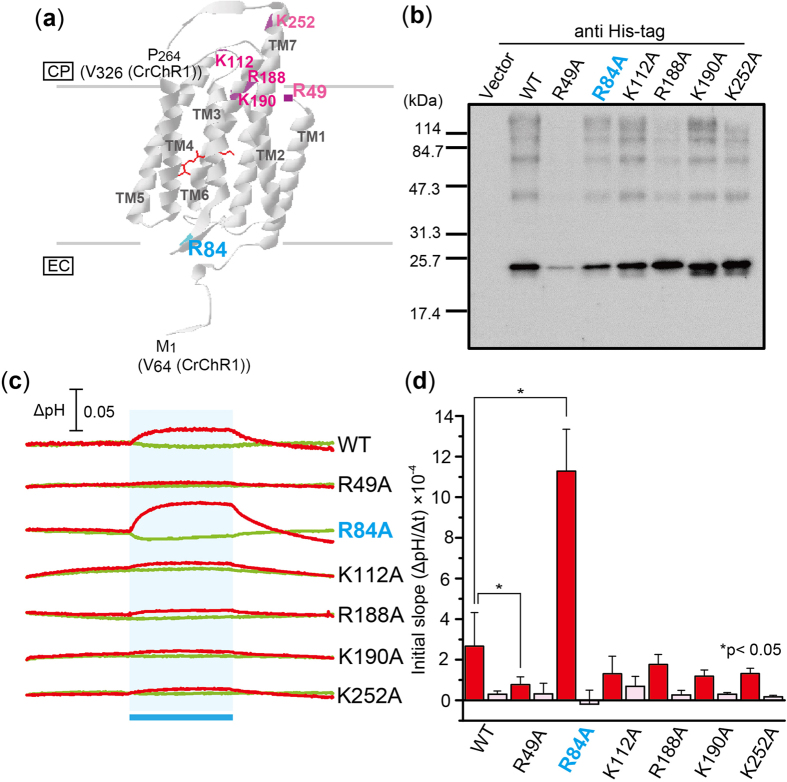
Figure 3. Effects of mutations on conserved basic amino acid residues in ACR2.
(a) Locations of mutations. The conserved basic amino acid residues in ACR2 are mapped on the crystal structure of a chimeric ChR (C1C2) from C. reinhardtii (PDB: 3UG9)21. CP and EC indicate the cytoplasmic and extracellular side, respectively. (b) Detection of protein expression of wild-type ACR2 and its mutants. These proteins were detected using an anti His-tag antibody. Cells harboring the pET22b vector plasmid alone were used as a negative control. (c) Light-induced pH changes of E. coli cells expressing ACR2 mutants in a solution containing 300 mM NaCl. The cell suspension was illuminated with blue light (480 ± 10 nm, ca. 9 mW/cm2) for 3 min (blue stripe). The red and green lines indicate pH changes in the presence or absence of 10 μM CCCP, respectively. (d) Comparison of the ion transport activity of the mutant proteins. The initial slope amplitudes of the light-induced pH changes of E. coli cell suspensions in 300 mM NaCl (red) or 100 mM Na2SO4 (pink) are plotted. Several independent experiments were averaged (n = 3–18). The error bars represent the standard deviation.