Fig. 5.
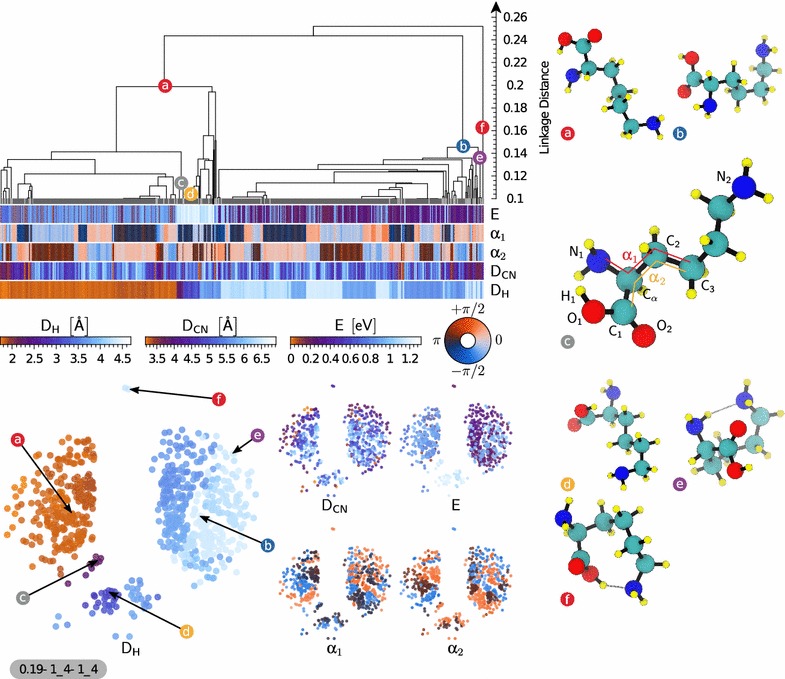
Representation of the similarity matrix corresponding to the lysine uncapped dataset using the agglomerative clustering algorithm (top) and the sketchmap algorithm (bottom, projection parameters shown following the scheme –A_B–a_b). A few representative structures (see Eq. 7) of interesting clusters are shown (right) and their corresponding position on the sketchmaps and dendrogram representation is highlighted. The five sketchmaps are colored according to the conformational energy, the distance between and the hydrogen in the carboxilic group (labelled ), the distance between and (labelled ), and the dihedral angles and which are respectively computed with the following atoms () and (). The dendrogram shows the clustering hierarchy of the structures of the dataset. Each structure is vertically aligned with its properties shown using color bars below the dendrogram. The dendrogram is cut at a linkage distance of 0.1 since structural properties are very similar below this threshold, and the clusters that are merged at this level are shown as thick gray bars separated by light-gray lines. Clusters composed of only one structure are drawn as a black line reaching the bottom of the dendrogram. The main structural motifs of the database are governed by the distance . The two main clusters a, b are agglomerated according to the orientation of and the oxygen atom it is bonded to with respect to which is well described by the distance . The sub-cluster e is composed of ‘outlier’ structures showing an H-bond between and an hydrogen of resulting in a folded side chain structural motif. Finally, the outlier cluster f contains a H-bond between the carboxy H and the side-chain , that can be seen as a precursor to the zwitterionic form
