Figure 5. Varying model parameters for integrase flipping and leaky expression.
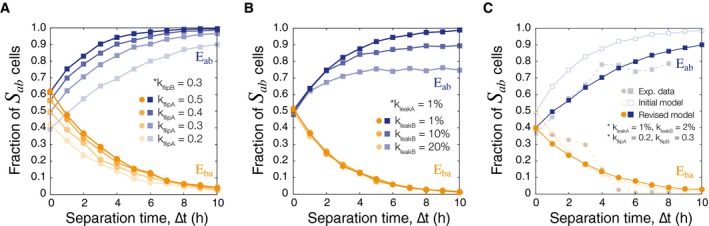
- As DNA flipping rates of intA () are decreased relative to , the population of S ab cells at ∆t = 0 h has a downward shift. Simulations are done with N = 3,000 trajectories/marker.
- Increasing the leaky expression of intB () changes the maximum threshold of cells that correctly identify S ab even at high ∆t. Leakiness is defined as a percentage of the induced integrase production rate ().
- The model was revised to more closely match the experimental data by constraining parameters for leaky expression and varying integrase flipping (N = 5,000). Mean squared error was calculated between the experimental data and the initial and revised models to find an optimized pair of values (Appendix Fig S13). The revised parameters are k flipA = 0.2 h−1, k flipB = 0.3 h−1, k leakA = 1% of k prodA(μm3 · h)−1, and k leakB = 2% of k prodB(μm3 · h)−1.
