Figure 2. Overview of the INDIGO approach to study drug interactions.
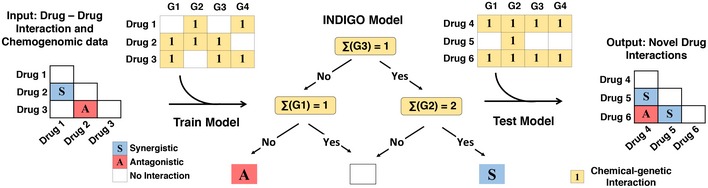
INDIGO takes in as input drug‐interaction data measuring synergistic and antagonistic interaction outcomes, and a compendium of chemogenomic profiles that identify gene deletions that enhance sensitivity to a compound of interest. INDIGO subsequently integrates the individual drug chemogenomic profiles using Boolean operations (sigma and delta scores) that capture similarity and dissimilarity (only sigma scores are shown for simplicity). INDIGO identifies top genomic predictors of drug synergy and antagonism using a random forest algorithm. The inferred model is then used to predict interaction outcomes for combinations of drugs with known chemogenomic profiles.
