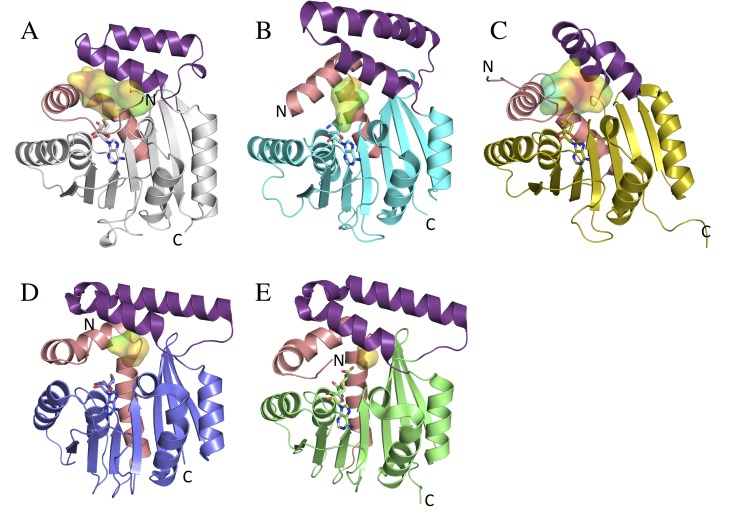
Fig 2. Overall structures of CouO and similar methyltransferases.
Monomers of A) CouO, B) Coq5 (PDB: 4obw), C) mmp1179 (PDB: 3dlc), D) YexO (PDB: 1im8) and CmoA (PDB: 4gek) are shown in ribbon representations. The cofactors (SAH in CouO, SAM in Coq5 and mmp1179, Cx-SAM in CmoA and SAI in YecO) are shown in sticks representations and the active site cavities are shown as surfaces. The cavity surfaces are colored according to their hydrophobicity (red-hydrophobic to blue-hydrophilic). The main differences are observed in the conformation of the cap-domain (violet) and in the position of the N-terminal part consisting of a loop and the first α-helix (salmon).