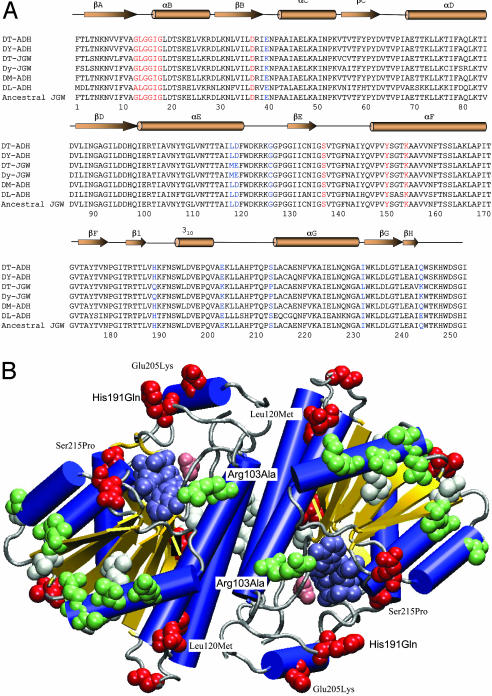
Fig. 2.
Structural analysis of JGW-specific amino acid substitutions. (A) Multiple sequence alignments of JGWs and related Drosophila ADHs (DT, D. teissieri; DY, D. yakuba; DM, D. melanogaster; and DL, D. lebanonensis). The parsimony inferred ancestral JGW sequence is unambiguous because the outgroup ADH sequences are highly similar to JGW. The secondary structure elements of DM-ADH (PDB ID code 1MG5), shown above the sequence alignments, include β-sheets, α-helices, and 310 helix. Red residues denote key positions in coenzyme binding. Blue residues denote nine positions with amino acid replacements accumulated early in the evolution of jgw. (B) Distribution of JGW-specific amino acid substitutions in the three-dimensional structure of ADH (blue helices, yellow sheets, and gray coils and turns) with bound acetate (pink van der Waals spheres) and NAD (steel van der Waals spheres). Of the nine amino acid replacements accumulated early in the evolution of JGW before the African species diverged (red van der Waals spheres), only two, His191Gln and Glu205Lys, lie in the flap region known to be important for substrate specificity. Two others, Ser215Pro and Leu120Met, contact residues within this region. All later replacements in the D. teissieri lineage (white van der Waals spheres) and in the D. yakuba lineage (green van der Waals spheres) lie outside the active site region.