Figure 1.
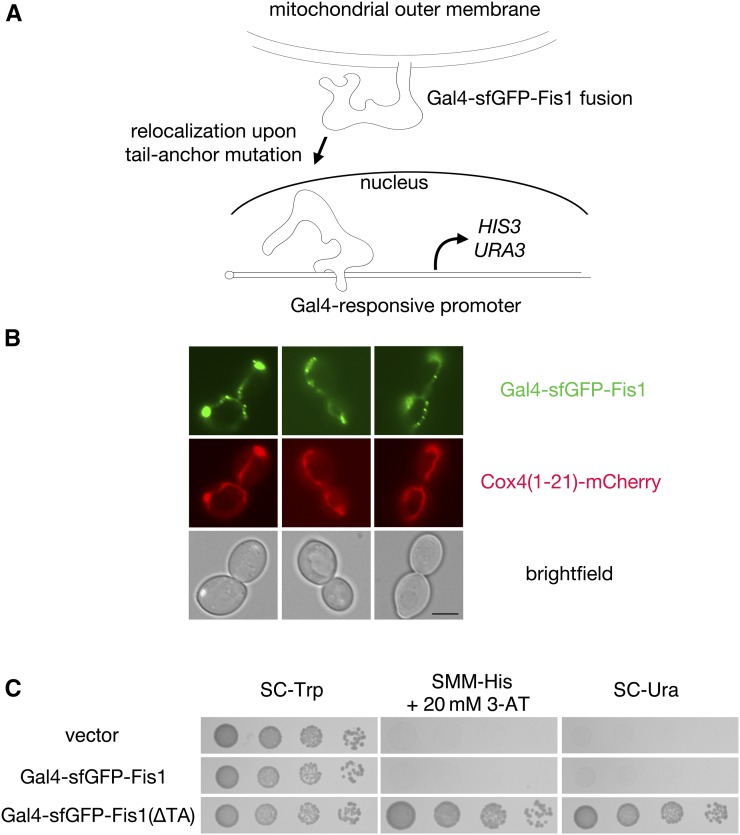
A genetic selection based on protein mislocalization allows recovery of mutations blocking Fis1p TA localization to mitochondria. (A) Scheme for selection of mutations preventing mitochondrial targeting of the Fis1p TA. Full-length Fis1p is fused to the transcription factor Gal4p. Upon failure of Fis1p to be localized to the mitochondrial OM, Gal4p may be free to translocate to the nucleus and activate the selectable markers HIS3 and URA3. (B) The Gal4–sfGFP–Fis1 fusion localizes to mitochondria. Strain CDD898 was transformed with plasmid b102, which overexpresses the Gal4–sfGFP–Fis1p construct used in this study. Mitochondria were visualized using mCherry fused to the Cox4 presequence expressed from plasmid pHS12–mCherry. Bar, 5 µm. (C) Removal of the TA from Gal4–sfGFP–Fis1p allows proliferation on medium requiring HIS3 activation or URA3 activation. Strain MaV203 expressing Gal4–sfGFP–Fis1p from plasmid b100 (WT TA), a variant lacking the Fis1p TA from plasmid b101 (∆TA), or harboring empty vector pKS1 was cultured in SC −Trp medium, then, following serial dilution, spotted to SC −Trp, SMM −His + 20 mM 3-AT, or SC −Ura and incubated for 2 days.