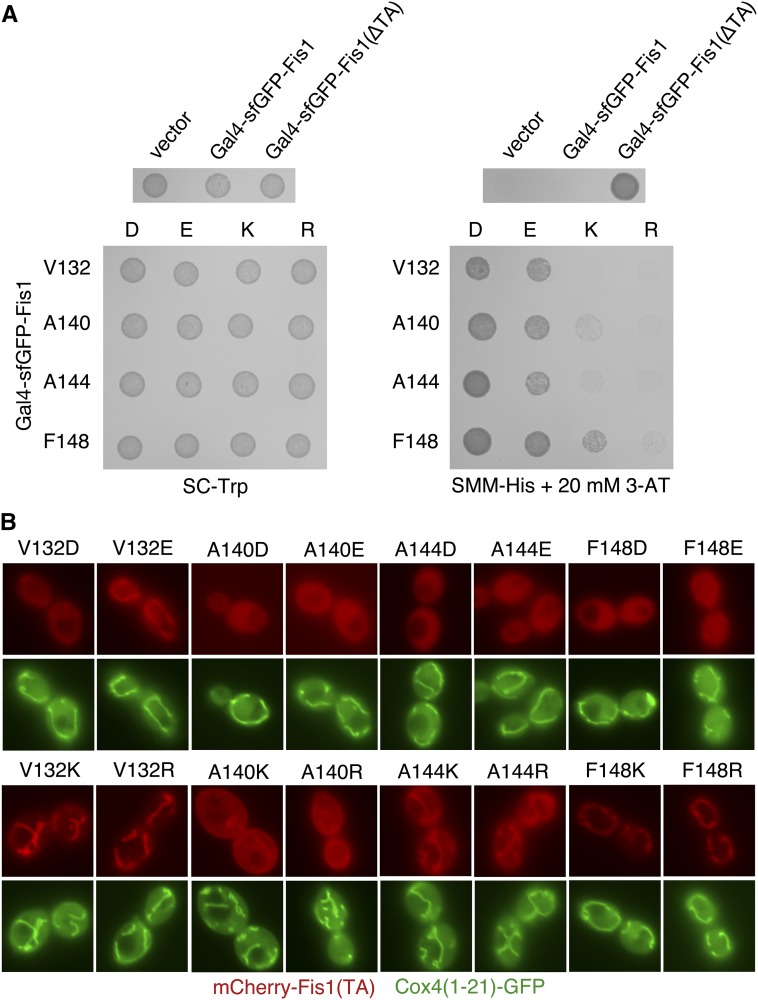
Figure 8.
Fis1p TA targeting is hindered to a greater extent by inclusion of negatively charged amino acids in the MAD than by positively charged amino acids. (A) Negative charges allow higher transcriptional activity than positive charges when placed at specific positions within the Gal4–sfGFP–Fis1p TA. Strain MaV203 was transformed with plasmids pKS1 (vector), b100 (Gal4–sfGFP–Fis1p), b101 [Gal4–sfGFP–Fis1p(∆TA)], or plasmids encoding the indicated charge replacements within the Fis1p TA (plasmids b173–b187 and b295). The resulting transformants were spotted to SC −Trp medium (1 day, 30°) or SMM −His + 20 mM 3-AT medium (2 days, 30°). (B) mCherry–Fis1(TA)p localization is disrupted more severely by negatively charged amino acids within the MAD than by positively charged amino acids. Strain CDD961 was transformed with plasmids (b192–b207) expressing mCherry linked to Fis1p TAs harboring the indicated substitutions. Cells were visualized as in Figure 2B. Bar, 5 µm.