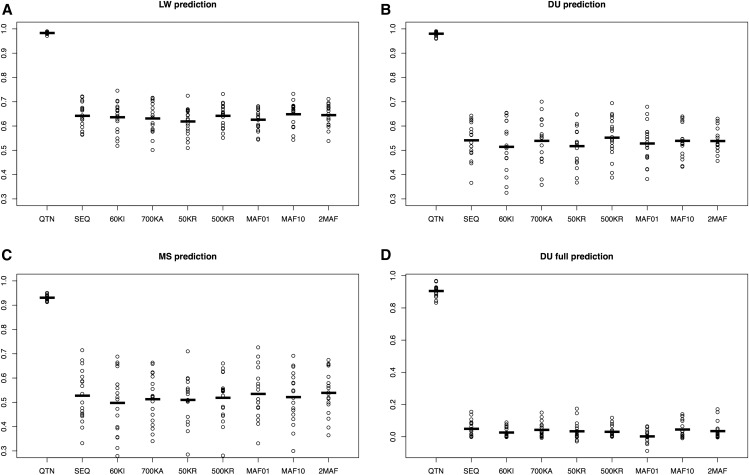
Figure 4.
Performance of different models across scenarios for the neutral architecture using GBLUP. (A) 50% of LW from the last generation were removed and predicted. (B) 50% of DU from the last generation removed. (C) 50% MS from the last generation removed. (D) All DU removed. The following models are shown: QTN, GRM computed with all QTNs; SEQ, all SNPs; 60KI, Illumina’s 60k array; 700KA, Affymetrix; 50KR, 50,000 random SNPs; 500KR, 500,000 random SNPs; MAF01, 1 million SNPs with MAF < 0.01; MAF10, 1 million SNPs with MAF > 0.1; and 2MAF, the model uses MAF01 and MAF10. Values reported are accuracies, defined as squared correlation between true and predicted genetic values. Dots correspond to each of the 10 replicates, horizontal bars represent the mean. DU, Duroc; GBLUP, genomic best linear unbiased prediction; GRM, genomic relationship matrix; LW, Large White; MAF, minimum allele frequency; MS, Meishan; QTN, quantitative trait nucleotide.