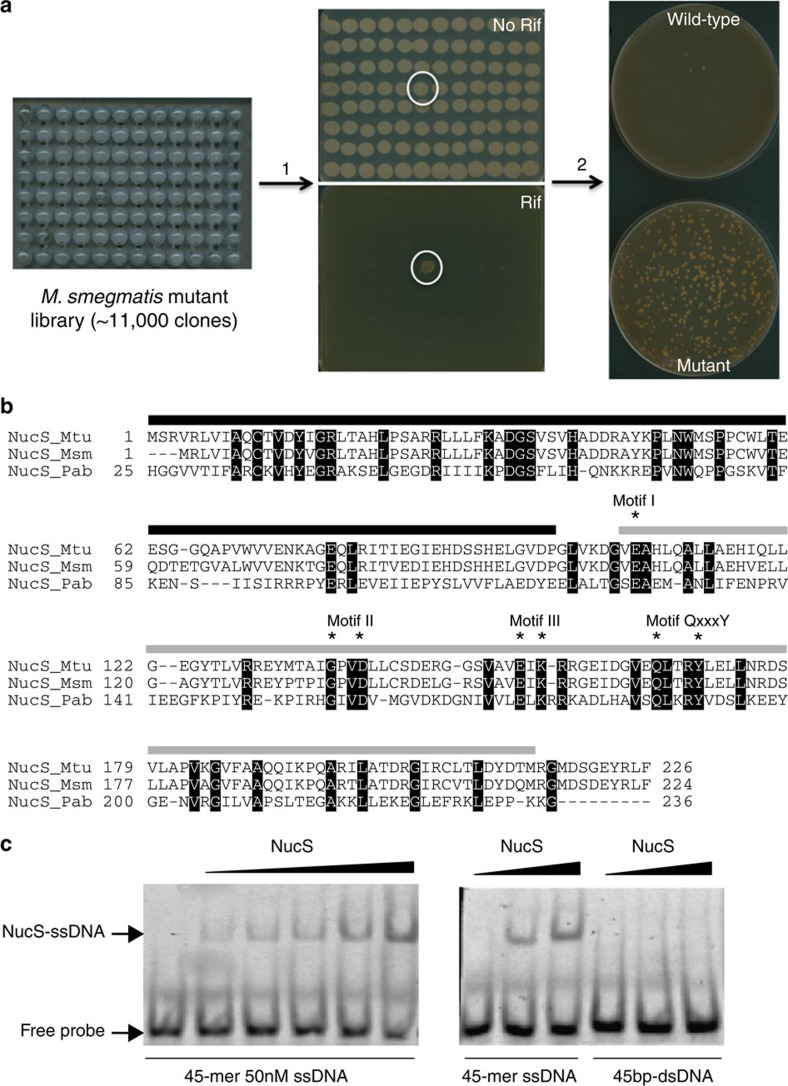
Figure 1. Identification and characterization of M. smegmatis NucS.
(a) Schematic representation of the process for identifying the nucS transposon mutant. ∼11,000 clones from the M. smegmatis insertion mutant library were replicated onto plates with (Rif) or without (No Rif) rifampicin (step 1). One single clone (circled) produced a high number of Rif-R colonies. After isolation and purification (step 2), the frequency of spontaneous Rif-R mutants (bottom plate) was checked and compared with that of the wild-type (upper plate), demonstrating its hypermutable phenotype. (b) Multiple sequence alignment of NucS sequences. M. tuberculosis (NucS_Mtu), M. smegmatis (NucS_Msm) and P. abyssi (NucS_Pab) sequences are from Uniprot (identifiers are P9WIY4, A0R1Z0 and Q9V2E8, respectively). Solid lines over the alignment indicate protein domains as defined previously for P. abyssi NucS11 (black, DNA-binding; grey, nuclease). Identical amino acid residues are shown in black. Catalytic residues required for nuclease activity in P. abyssi NucS11 are labelled with asterisks. Nuclease motifs of P. abyssi NucS are also indicated11. (c) DNA-binding activity of NucS. In a gel-based EMSA, purified NucS protein (1–16 μM) is capable of binding to 45-mer ssDNA (50 nM) (left side) but not to 45-bp dsDNA (50 nM) (right side). The arrow indicates the position of the DNA–NucS complex.