Figure 5.
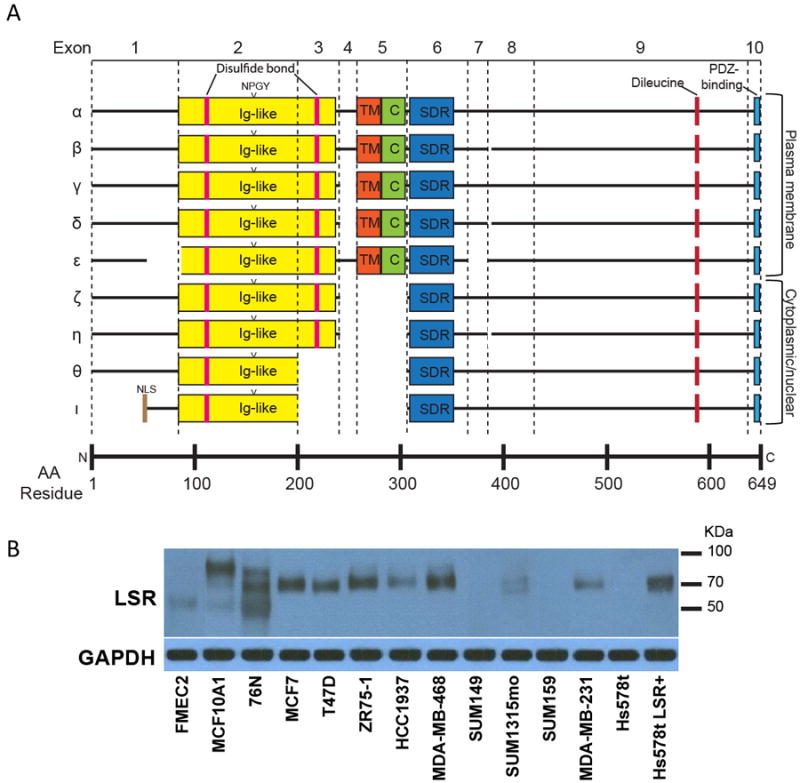
Potential LSR transcript variants. A, LSR transcripts α-ι were identified from online databases and are shown with putative structural and protein-binding domains. Most LSR transcript variants appear to be the result of alternative splicing of particular exons. Ig-like – immunoglobulin-like; NLS – nuclear localization sequence; TM – transmembrane; C – cysteine-rich; SDR – short-chain dehydrogenase/reductase. B, Fourteen cell lines were examined for the expression of LSR (top panel) and a GAPDH loading control (bottom panel) by western blot analysis. Different cell lines expressed LSR at varying levels and at different molecular weights when using a polyclonal antibody that recognizes all predicted isoforms of LSR. Immunoblots are representative of a minimum of three independent experiments.
