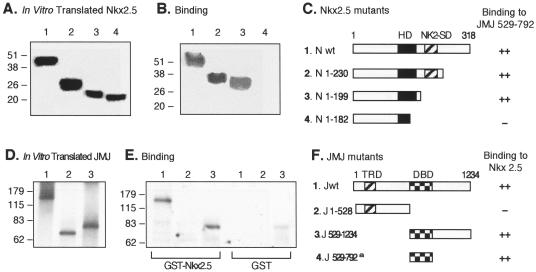
FIG. 5.
Mapping of the protein-protein interaction domains in Nkx2.5 and JMJ. Various 35S-labeled Nkx2.5 mutants were prepared by using an in vitro transcription and translation kit (Promega) and confirmed by SDS-PAGE and autoradiography (A). Equal amounts of 35S-Nkx2.5 were incubated with approximately 1 μg of GST-JMJ 529-792 fusion proteins coupled to agarose beads followed by extensive washing and SDS-PAGE (B). A diagram of protein structures for the Nkx2.5 mutants and a summary of the physical interaction with JMJ are shown (C). To perform reciprocal experiments, various 35S-labeled JMJ mutants (D) were incubated with approximately 1 μg of GST-Nkx2.5 beads or GST beads, as indicated, followed by SDS-PAGE (E). A diagram of protein structures for JMJ mutants and a summary of binding to Nkx2.5 are shown (F); the a indicates the binding activity of GST-JMJ 529-792 to 35S-Nkx2.5 as shown in panels B and C. The homeodomain of Nkx2.5 (HD) and the Nkx2-specific domain (NK2-SD) consist of aa 137 to 196 and 210 to 226, respectively (27). The transcriptional repression domain (TRD) and DBD of JMJ consist of aa 131 to 222 and 529 to 792, respectively. Association affinity: ++, strong interaction; +, moderate interaction; −, no interaction. N, Nkx2.5; J, JMJ; wt, wild type.