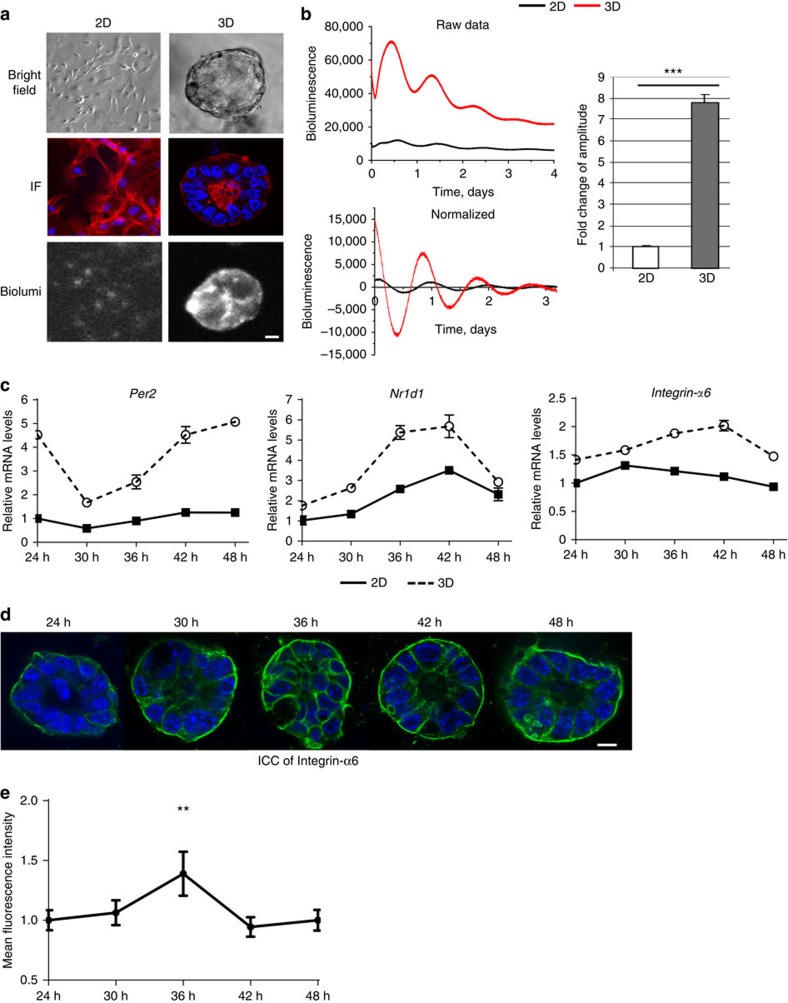
Figure 5. The extracellular microenvironment regulates mammary clock.
(a) Brightfield: primary MECs form monolayers in 2D collagen (2D) or acini structures in 3D Matrigel (3D), shown by phase contrast microscopy. N=3 animals, scale bar=20 μm. IF: red, β-Actin staining; blue, DAPI. Biolum: Similar cultures shown by bioluminescence staining of PER2::Luc. (b) Bioluminescence recordings and quantification of PER2::Luc activity from the same number of MECs cultured either on 2D or 3D. Student's t-test, mean±s.e.m., ***P<0.001, n=6 animals. The fold amplitude in difference in activity is shown on the right. (c) The expression patterns of endogenous clock genes (Per2 and Nr1d1), and clock target gene (for example, α6-integrin) in MECs cultured on either 2D or 3D, n=4 animals. The mRNA levels were normalized to GAPDH and then to the 2D condition at 24 h. (d) Representative IF staining (blue, DAPI; green, integrin-α6) and (e) semi-quantification of integrin-α6 levels in MECs from WT mice cultured in 3D on Matrigel. Scale bar=20 μm. **P<0.01 for time-dependent expression, one-way ANOVA, n=3 animals.